+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6j56 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
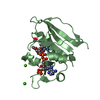
| Title | Crystal structure of Myosin VI CBD in complex with Tom1 MBM | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | PROTEIN BINDING / Myosin VI / Tom1 / complex / autophagy | |||||||||
| Function / homology |  Function and homology information Function and homology informationmyosin VI binding / substrate localization to autophagosome / regulation of endosome organization / regulation of secretion / minus-end directed microfilament motor activity / unconventional myosin complex / clathrin heavy chain binding / phosphatidylinositol-5-phosphate binding / autophagosome-lysosome fusion / inner ear auditory receptor cell differentiation ...myosin VI binding / substrate localization to autophagosome / regulation of endosome organization / regulation of secretion / minus-end directed microfilament motor activity / unconventional myosin complex / clathrin heavy chain binding / phosphatidylinositol-5-phosphate binding / autophagosome-lysosome fusion / inner ear auditory receptor cell differentiation / actin filament-based movement / clathrin-coated vesicle membrane / Gap junction degradation / Trafficking of AMPA receptors / positive regulation of autophagosome maturation / RHOBTB1 GTPase cycle / endosomal transport / inner ear morphogenesis / azurophil granule membrane / clathrin binding / microfilament motor activity / filamentous actin / microvillus / RHOU GTPase cycle / cytoskeletal motor activity / polyubiquitin modification-dependent protein binding / endocytic vesicle / RHOBTB2 GTPase cycle / specific granule membrane / ruffle / clathrin-coated pit / autophagosome / actin filament organization / ubiquitin binding / actin filament / filopodium / intracellular protein transport / DNA damage response, signal transduction by p53 class mediator / sensory perception of sound / ADP binding / ruffle membrane / endocytosis / apical part of cell / actin filament binding / intracellular protein localization / actin cytoskeleton / protein transport / actin binding / cytoplasmic vesicle / early endosome membrane / cell cortex / nuclear membrane / early endosome / calmodulin binding / endosome / endosome membrane / response to xenobiotic stimulus / lysosomal membrane / Neutrophil degranulation / perinuclear region of cytoplasm / Golgi apparatus / signal transduction / extracellular exosome / nucleoplasm / ATP binding / identical protein binding / nucleus / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.798 Å MOLECULAR REPLACEMENT / Resolution: 1.798 Å | |||||||||
 Authors Authors | Hu, S. / Pan, L. | |||||||||
| Funding support |  China, 2items China, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Structure of Myosin VI/Tom1 complex reveals a cargo recognition mode of Myosin VI for tethering. Authors: Hu, S. / Guo, Y. / Wang, Y. / Li, Y. / Fu, T. / Zhou, Z. / Wang, Y. / Liu, J. / Pan, L. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6j56.cif.gz 6j56.cif.gz | 135.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6j56.ent.gz pdb6j56.ent.gz | 104.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6j56.json.gz 6j56.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/j5/6j56 https://data.pdbj.org/pub/pdb/validation_reports/j5/6j56 ftp://data.pdbj.org/pub/pdb/validation_reports/j5/6j56 ftp://data.pdbj.org/pub/pdb/validation_reports/j5/6j56 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3h8dS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 15370.664 Da / Num. of mol.: 2 / Fragment: CBD domain Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: MYO6, KIAA0389 Homo sapiens (human) / Gene: MYO6, KIAA0389Production host:  References: UniProt: Q9UM54 #2: Protein/peptide | Mass: 2971.303 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TOM1 Homo sapiens (human) / Gene: TOM1Production host:  References: UniProt: O60784 #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.03 Å3/Da / Density % sol: 39.39 % |
|---|---|
| Crystal grow | Temperature: 289 K / Method: vapor diffusion, sitting drop Details: PEG 4000, sodium acetate trihydrate, TRIS hydrochloride |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRF SSRF  / Beamline: BL19U1 / Wavelength: 0.97774 Å / Beamline: BL19U1 / Wavelength: 0.97774 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Dec 1, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97774 Å / Relative weight: 1 |
| Reflection | Resolution: 1.798→50 Å / Num. obs: 26915 / % possible obs: 98.5 % / Redundancy: 6.6 % / Rmerge(I) obs: 0.056 / Net I/σ(I): 28.14 |
| Reflection shell | Resolution: 1.8→1.83 Å / Redundancy: 6.2 % / Num. unique obs: 1342 / CC1/2: 0.976 / Rpim(I) all: 0.149 / % possible all: 98.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3H8D Resolution: 1.798→37.438 Å / SU ML: 0.17 / Cross valid method: NONE / σ(F): 1.34 / Phase error: 22.55
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.798→37.438 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj











