| Entry | Database: PDB / ID: 6gy4
|
|---|
| Title | Crystal structure of the N-terminal KH domain of human BICC1 |
|---|
 Components Components | Protein bicaudal C homolog 1 |
|---|
 Keywords Keywords | RNA BINDING PROTEIN / Bicaudal / RNA binding / Transcription / KH domain |
|---|
| Function / homology |  Function and homology information Function and homology information
determination of left/right symmetry / kidney development / negative regulation of canonical Wnt signaling pathway / heart development / RNA binding / cytoplasmSimilarity search - Function BICC1, SAM domain / : / : / : / : / Protein bicaudal C homolog 1, KH-like domain / BICC1, first type I KH domain / KH domain / K Homology domain, type 1 / SAM domain (Sterile alpha motif) ...BICC1, SAM domain / : / : / : / : / Protein bicaudal C homolog 1, KH-like domain / BICC1, first type I KH domain / KH domain / K Homology domain, type 1 / SAM domain (Sterile alpha motif) / Type-1 KH domain profile. / K Homology domain, type 1 superfamily / SAM domain profile. / Sterile alpha motif. / Sterile alpha motif domain / Sterile alpha motif/pointed domain superfamily / K Homology domain / K homology RNA-binding domainSimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.986 Å MOLECULAR REPLACEMENT / Resolution: 1.986 Å |
|---|
 Authors Authors | Newman, J.A. / Katis, V.L. / Shrestha, L. / Burgess-Brown, N. / von Delft, F. / Arrowsmith, C.H. / Edwards, A. / Bountra, C. / Gileadi, O. |
|---|
| Funding support |  United Kingdom, 1items United Kingdom, 1items | Organization | Grant number | Country |
|---|
| Wellcome Trust | |  United Kingdom United Kingdom |
|
|---|
 Citation Citation |  Journal: To Be Published Journal: To Be Published
Title: Crystal structure of the N-terminal KH domain of human BICC1
Authors: Newman, J.A. / Katis, V.L. / Shrestha, L. / Burgess-Brown, N. / von Delft, F. / Arrowsmith, C.H. / Edwards, A. / Bountra, C. / Gileadi, O. |
|---|
| History | | Deposition | Jun 28, 2018 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Jul 18, 2018 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jan 17, 2024 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.986 Å
MOLECULAR REPLACEMENT / Resolution: 1.986 Å  Authors
Authors United Kingdom, 1items
United Kingdom, 1items  Citation
Citation Journal: To Be Published
Journal: To Be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6gy4.cif.gz
6gy4.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6gy4.ent.gz
pdb6gy4.ent.gz PDB format
PDB format 6gy4.json.gz
6gy4.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/gy/6gy4
https://data.pdbj.org/pub/pdb/validation_reports/gy/6gy4 ftp://data.pdbj.org/pub/pdb/validation_reports/gy/6gy4
ftp://data.pdbj.org/pub/pdb/validation_reports/gy/6gy4
 Links
Links Assembly
Assembly
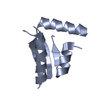



 Components
Components Homo sapiens (human) / Gene: BICC1 / Production host:
Homo sapiens (human) / Gene: BICC1 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I04-1 / Wavelength: 0.9212 Å
/ Beamline: I04-1 / Wavelength: 0.9212 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj






