+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ge1 | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Solution structure of the r(UGGUGGU)4 RNA quadruplex | |||||||||||||||||||||||||||||||
 Components Components | RNA (5'-R(* Keywords KeywordsRNA / quadruplex / tetramolecular / parallel stranded / U-tetrad | Function / homology | RNA |  Function and homology information Function and homology informationBiological species | synthetic construct (others) | Method | SOLUTION NMR / molecular dynamics / simulated annealing |  Authors AuthorsAndralojc, W. / Gdaniec, Z. | Funding support | |  Poland, 2items Poland, 2items
 Citation Citation Journal: RNA / Year: 2019 Journal: RNA / Year: 2019Title: Unraveling the structural basis for the exceptional stability of RNA G-quadruplexes capped by a uridine tetrad at the 3' terminus. Authors: Andralojc, W. / Malgowska, M. / Sarzynska, J. / Pasternak, K. / Szpotkowski, K. / Kierzek, R. / Gdaniec, Z. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ge1.cif.gz 6ge1.cif.gz | 260.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ge1.ent.gz pdb6ge1.ent.gz | 221.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ge1.json.gz 6ge1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6ge1_validation.pdf.gz 6ge1_validation.pdf.gz | 532.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6ge1_full_validation.pdf.gz 6ge1_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  6ge1_validation.xml.gz 6ge1_validation.xml.gz | 54 KB | Display | |
| Data in CIF |  6ge1_validation.cif.gz 6ge1_validation.cif.gz | 53.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ge/6ge1 https://data.pdbj.org/pub/pdb/validation_reports/ge/6ge1 ftp://data.pdbj.org/pub/pdb/validation_reports/ge/6ge1 ftp://data.pdbj.org/pub/pdb/validation_reports/ge/6ge1 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 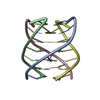
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 2254.363 Da / Num. of mol.: 4 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||
| Sample conditions |
|
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE III / Manufacturer: Bruker / Model: AVANCE III / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement |
| |||||||||||||||
| NMR representative | Selection criteria: lowest energy | |||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 400 / Conformers submitted total number: 15 |
 Movie
Movie Controller
Controller










 PDBj
PDBj SAXS
SAXS