[English] 日本語
 Yorodumi
Yorodumi- PDB-6fqn: Carbamylated T. californica acetylcholineterase bound to uncharge... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6fqn | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Carbamylated T. californica acetylcholineterase bound to uncharged hybrid reactivator 2 | |||||||||
 Components Components | Acetylcholinesterase | |||||||||
 Keywords Keywords | HYDROLASE / Acetylcholinesterase Hybrid reactivators Organophosphate inhibition Structure-based optimization | |||||||||
| Function / homology |  Function and homology information Function and homology informationacetylcholine catabolic process in synaptic cleft / acetylcholinesterase / choline metabolic process / acetylcholinesterase activity / synaptic cleft / side of membrane / synapse / extracellular space / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.30002662121 Å MOLECULAR REPLACEMENT / Resolution: 2.30002662121 Å | |||||||||
 Authors Authors | De la Mora, E. / Santoni, G. / de Souza, J. / Sussman, J. / Silman, I. / Baati, R. / Weik, M. / Nachon, F. | |||||||||
| Funding support |  France, 2items France, 2items
| |||||||||
 Citation Citation |  Journal: J. Med. Chem. / Year: 2018 Journal: J. Med. Chem. / Year: 2018Title: Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents. Authors: Santoni, G. / de Sousa, J. / de la Mora, E. / Dias, J. / Jean, L. / Sussman, J.L. / Silman, I. / Renard, P.Y. / Brown, R.C.D. / Weik, M. / Baati, R. / Nachon, F. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6fqn.cif.gz 6fqn.cif.gz | 138.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6fqn.ent.gz pdb6fqn.ent.gz | 97.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6fqn.json.gz 6fqn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fq/6fqn https://data.pdbj.org/pub/pdb/validation_reports/fq/6fqn ftp://data.pdbj.org/pub/pdb/validation_reports/fq/6fqn ftp://data.pdbj.org/pub/pdb/validation_reports/fq/6fqn | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6fldC  6g17C  6g4mC  6g4nC  6g4oC  6g4pC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein / Sugars , 2 types, 4 molecules A

| #1: Protein | Mass: 60279.078 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: ache Production host:  References: UniProt: P04058, acetylcholinesterase |
|---|---|
| #3: Sugar |
-Non-polymers , 5 types, 173 molecules 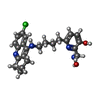








| #2: Chemical | ChemComp-E2W / | ||||||
|---|---|---|---|---|---|---|---|
| #4: Chemical | | #5: Chemical | ChemComp-CL / #6: Chemical | ChemComp-P6G / | #7: Water | ChemComp-HOH / | |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.24 Å3/Da / Density % sol: 70.98 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion / Details: MES 100 mM pH 5.4 PEG 200 28% |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-2 / Wavelength: 0.873 Å / Beamline: ID23-2 / Wavelength: 0.873 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Oct 31, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.873 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→39.98 Å / Num. obs: 45946 / % possible obs: 99.72 % / Observed criterion σ(I): 2 / Redundancy: 6.9 % / Biso Wilson estimate: 41.15 Å2 / CC1/2: 0.998 / Rmerge(I) obs: 0.106 / Rpim(I) all: 0.042 / Rrim(I) all: 0.115 / Net I/σ(I): 11.3 |
| Reflection shell | Resolution: 2.3→2.38 Å / Redundancy: 6.7 % / Rmerge(I) obs: 0.691 / Mean I/σ(I) obs: 2.33 / Num. unique obs: 4513 / CC1/2: 0.812 / Rpim(I) all: 0.285 / Rrim(I) all: 0.7493 / % possible all: 99.73 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.30002662121→39.9721095433 Å / SU ML: 0.336235589523 / Cross valid method: FREE R-VALUE / σ(F): 1.35120167119 / Phase error: 28.7743616241 MOLECULAR REPLACEMENT / Resolution: 2.30002662121→39.9721095433 Å / SU ML: 0.336235589523 / Cross valid method: FREE R-VALUE / σ(F): 1.35120167119 / Phase error: 28.7743616241
| ||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||
| Displacement parameters | Biso mean: 51.7376603806 Å2 | ||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.30002662121→39.9721095433 Å
| ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj





