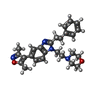
登録情報 データベース : PDB / ID : 6dmkタイトル A multiconformer ligand model of an isoxazolyl-benzimidazole ligand bound to the bromodomain of human CREBBP CREB-binding protein キーワード / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト)手法 / / 解像度 : 1.66 Å データ登録者 Hudson, B.M. / van Zundert, G. / Keedy, D.A. / Fonseca, R. / Heliou, A. / Suresh, P. / Borrelli, K. / Day, T. / Fraser, J.S. / van den Bedem, H. ジャーナル : J. Med. Chem. / 年 : 2018タイトル : qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.著者 : van Zundert, G.C.P. / Hudson, B.M. / de Oliveira, S.H.P. / Keedy, D.A. / Fonseca, R. / Heliou, A. / Suresh, P. / Borrelli, K. / Day, T. / Fraser, J.S. / van den Bedem, H. 履歴 登録 2018年6月5日 登録サイト / 処理サイト 改定 1.0 2018年12月19日 Provider / タイプ 改定 1.1 2019年4月24日 Group / Database references / カテゴリ / citation_authorItem _citation.journal_volume / _citation.page_first ... _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.title / _citation_author.identifier_ORCID / _citation_author.name 改定 1.2 2024年3月13日 Group / Database references / カテゴリ / chem_comp_bond / database_2Item / _database_2.pdbx_database_accession改定 1.3 2024年5月1日 Group / カテゴリ / Item
すべて表示 表示を減らす Remark 0 THIS ENTRY 6DMK REFLECTS AN ALTERNATIVE MODELING OF THE ORIGINAL DATA IN 4NR5, DETERMINED BY P. ... THIS ENTRY 6DMK REFLECTS AN ALTERNATIVE MODELING OF THE ORIGINAL DATA IN 4NR5, DETERMINED BY P.FILIPPAKOPOULOS,S.PICAUD,I.FELLETAR,D.HAY,O.FEDOROV,S.MARTIN,A.W.PIKE,F.VON DELFT,P.BRENNAN,C.H.ARROWSMITH,A.M.EDWARDS,C.BOUNTRA,S.KNAPP,STRUCTURAL GENOMICS CONSORTIUM (SGC)
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) X線回折 /
X線回折 /  シンクロトロン / 解像度: 1.66 Å
シンクロトロン / 解像度: 1.66 Å  データ登録者
データ登録者 引用
引用 ジャーナル: J. Med. Chem. / 年: 2018
ジャーナル: J. Med. Chem. / 年: 2018 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 6dmk.cif.gz
6dmk.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb6dmk.ent.gz
pdb6dmk.ent.gz PDB形式
PDB形式 6dmk.json.gz
6dmk.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 6dmk_validation.pdf.gz
6dmk_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 6dmk_full_validation.pdf.gz
6dmk_full_validation.pdf.gz 6dmk_validation.xml.gz
6dmk_validation.xml.gz 6dmk_validation.cif.gz
6dmk_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/dm/6dmk
https://data.pdbj.org/pub/pdb/validation_reports/dm/6dmk ftp://data.pdbj.org/pub/pdb/validation_reports/dm/6dmk
ftp://data.pdbj.org/pub/pdb/validation_reports/dm/6dmk リンク
リンク 集合体
集合体
 要素
要素 Homo sapiens (ヒト) / 遺伝子: CREBBP, CBP / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: CREBBP, CBP / 発現宿主: 
 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  Diamond
Diamond  / ビームライン: I24 / 波長: 0.9686 Å
/ ビームライン: I24 / 波長: 0.9686 Å 解析
解析 ムービー
ムービー コントローラー
コントローラー


















 PDBj
PDBj