[English] 日本語
 Yorodumi
Yorodumi- PDB-6dm6: Structure of DNA polymerase III subunit beta from Rickettsia cono... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6dm6 | ||||||
|---|---|---|---|---|---|---|---|
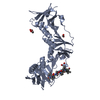
| Title | Structure of DNA polymerase III subunit beta from Rickettsia conorii in complex with a natural product | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSFERASE / SSGCID / dnaN / DNA polymerase III subunit beta / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease | ||||||
| Function / homology |  Function and homology information Function and homology informationDNA polymerase III complex / DNA strand elongation involved in DNA replication / 3'-5' exonuclease activity / DNA-directed DNA polymerase activity / DNA binding / cytoplasm Similarity search - Function | ||||||
| Biological species |  Rickettsia conorii (bacteria) Rickettsia conorii (bacteria) Streptomyces caelicus (bacteria) Streptomyces caelicus (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.25 Å MOLECULAR REPLACEMENT / Resolution: 2.25 Å | ||||||
 Authors Authors | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | ||||||
 Citation Citation |  Journal: to be published Journal: to be publishedTitle: Structure of DNA polymerase III subunit beta from Rickettsia conorii in complex with a natural product Authors: Conrady, D.G. / Abendroth, J. / Higgins, T.W. / Lorimer, D.D. / Horanyi, P.S. / Edwards, T.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6dm6.cif.gz 6dm6.cif.gz | 320.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6dm6.ent.gz pdb6dm6.ent.gz | 256.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6dm6.json.gz 6dm6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dm/6dm6 https://data.pdbj.org/pub/pdb/validation_reports/dm/6dm6 ftp://data.pdbj.org/pub/pdb/validation_reports/dm/6dm6 ftp://data.pdbj.org/pub/pdb/validation_reports/dm/6dm6 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6dlkS S: Starting model for refinement |
|---|---|
| Similar structure data | |
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments:
|
 Movie
Movie Controller
Controller











 PDBj
PDBj