| Entry | Database: PDB / ID: 6bz1
|
|---|
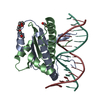
| Title | MEF2 Chimera D83V mutant/DNA complex |
|---|
 Components Components | - DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3')
- DNA (5'-D(P*TP*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3')
- MEF2 CHIMERA
|
|---|
 Keywords Keywords | TRANSCRIPTION/DNA / MEF2 / Transcription Factor / D83V mutant / conformation switch / TRANSCRIPTION-DNA complex |
|---|
| Function / homology |  Function and homology information Function and homology information
ERK5 cascade / ventricular cardiac myofibril assembly / mitochondrion distribution / cardiac conduction / : / muscle organ development / Myogenesis / dendrite morphogenesis / positive regulation of cardiac muscle hypertrophy / histone acetyltransferase binding ...ERK5 cascade / ventricular cardiac myofibril assembly / mitochondrion distribution / cardiac conduction / : / muscle organ development / Myogenesis / dendrite morphogenesis / positive regulation of cardiac muscle hypertrophy / histone acetyltransferase binding / SMAD binding / ERK/MAPK targets / cellular response to calcium ion / positive regulation of D-glucose import across plasma membrane / RNA polymerase II transcription regulatory region sequence-specific DNA binding / histone deacetylase binding / sequence-specific double-stranded DNA binding / cell junction / MAPK cascade / heart development / DNA-binding transcription activator activity, RNA polymerase II-specific / transcription regulator complex / sequence-specific DNA binding / DNA-binding transcription factor binding / RNA polymerase II-specific DNA-binding transcription factor binding / DNA-binding transcription factor activity, RNA polymerase II-specific / cell differentiation / protein dimerization activity / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / protein heterodimerization activity / apoptotic process / DNA-templated transcription / chromatin binding / positive regulation of gene expression / protein kinase binding / chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / nucleoplasm / nucleus / cytosolSimilarity search - Function SRF-like / Transcription factor, MADS-box / Holliday junction regulator protein family C-terminal / Holliday junction regulator protein family C-terminal repeat / MADS MEF2-like / Transcription factor, MADS-box / Transcription factor, MADS-box superfamily / SRF-type transcription factor (DNA-binding and dimerisation domain) / MADS-box domain signature. / MADS-box domain profile. ...SRF-like / Transcription factor, MADS-box / Holliday junction regulator protein family C-terminal / Holliday junction regulator protein family C-terminal repeat / MADS MEF2-like / Transcription factor, MADS-box / Transcription factor, MADS-box superfamily / SRF-type transcription factor (DNA-binding and dimerisation domain) / MADS-box domain signature. / MADS-box domain profile. / MADS / 3-Layer(aba) Sandwich / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.97 Å MOLECULAR REPLACEMENT / Resolution: 2.97 Å |
|---|
 Authors Authors | Lei, X. / Chen, L. |
|---|
| Funding support |  United States, 1items United States, 1items | Organization | Grant number | Country |
|---|
| National Institutes of Health/National Human Genome Research Institute (NIH/NHGRI) | |  United States United States |
|
|---|
 Citation Citation |  Journal: J. Mol. Biol. / Year: 2018 Journal: J. Mol. Biol. / Year: 2018
Title: The Cancer Mutation D83V Induces an alpha-Helix to beta-Strand Conformation Switch in MEF2B.
Authors: Lei, X. / Kou, Y. / Fu, Y. / Rajashekar, N. / Shi, H. / Wu, F. / Xu, J. / Luo, Y. / Chen, L. |
|---|
| History | | Deposition | Dec 21, 2017 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Feb 7, 2018 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Feb 14, 2018 | Group: Derived calculations
Category: pdbx_struct_assembly_gen / pdbx_struct_assembly_prop
Item: _pdbx_struct_assembly_gen.asym_id_list / _pdbx_struct_assembly_prop.value |
|---|
| Revision 1.2 | Mar 7, 2018 | Group: Database references / Category: citation / citation_author
Item: _citation.country / _citation.journal_abbrev ..._citation.country / _citation.journal_abbrev / _citation.journal_id_ASTM / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year |
|---|
| Revision 1.3 | Apr 11, 2018 | Group: Data collection / Database references / Category: citation
Item: _citation.journal_volume / _citation.page_first ..._citation.journal_volume / _citation.page_first / _citation.page_last / _citation.title |
|---|
| Revision 1.4 | Dec 18, 2019 | Group: Author supporting evidence / Category: pdbx_audit_support / Item: _pdbx_audit_support.funding_organization |
|---|
| Revision 1.5 | Mar 13, 2024 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / struct_ncs_dom_lim
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.97 Å
MOLECULAR REPLACEMENT / Resolution: 2.97 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: J. Mol. Biol. / Year: 2018
Journal: J. Mol. Biol. / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6bz1.cif.gz
6bz1.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6bz1.ent.gz
pdb6bz1.ent.gz PDB format
PDB format 6bz1.json.gz
6bz1.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/bz/6bz1
https://data.pdbj.org/pub/pdb/validation_reports/bz/6bz1 ftp://data.pdbj.org/pub/pdb/validation_reports/bz/6bz1
ftp://data.pdbj.org/pub/pdb/validation_reports/bz/6bz1 Links
Links Assembly
Assembly


 Movie
Movie Controller
Controller











 PDBj
PDBj






































