[English] 日本語
 Yorodumi
Yorodumi- PDB-5wsb: Pyruvate kinase (PYK) from Mycobacterium tuberculosis in complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5wsb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pyruvate kinase (PYK) from Mycobacterium tuberculosis in complex with Oxalate, allosteric activators AMP and Glucose 6-Phosphate | |||||||||
 Components Components | Pyruvate kinase | |||||||||
 Keywords Keywords | TRANSFERASE / pyruvate kinase / glycolysis / tetramer / allostery / synergism / phospho transferase | |||||||||
| Function / homology |  Function and homology information Function and homology informationpyruvate kinase / pyruvate kinase activity / potassium ion binding / glycolytic process / kinase activity / magnesium ion binding / ATP binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.25 Å MOLECULAR REPLACEMENT / Resolution: 2.25 Å | |||||||||
 Authors Authors | Zhong, W. / Cai, Q. / El Sahili, A. / Lescar, J. / Dedon, P.C. | |||||||||
| Funding support |  Singapore, 2items Singapore, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Allosteric pyruvate kinase-based "logic gate" synergistically senses energy and sugar levels in Mycobacterium tuberculosis. Authors: Zhong, W. / Cui, L. / Goh, B.C. / Cai, Q. / Ho, P. / Chionh, Y.H. / Yuan, M. / Sahili, A.E. / Fothergill-Gilmore, L.A. / Walkinshaw, M.D. / Lescar, J. / Dedon, P.C. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5wsb.cif.gz 5wsb.cif.gz | 704.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5wsb.ent.gz pdb5wsb.ent.gz | 584 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5wsb.json.gz 5wsb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  5wsb_validation.pdf.gz 5wsb_validation.pdf.gz | 2.5 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  5wsb_full_validation.pdf.gz 5wsb_full_validation.pdf.gz | 2.5 MB | Display | |
| Data in XML |  5wsb_validation.xml.gz 5wsb_validation.xml.gz | 67.4 KB | Display | |
| Data in CIF |  5wsb_validation.cif.gz 5wsb_validation.cif.gz | 94.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ws/5wsb https://data.pdbj.org/pub/pdb/validation_reports/ws/5wsb ftp://data.pdbj.org/pub/pdb/validation_reports/ws/5wsb ftp://data.pdbj.org/pub/pdb/validation_reports/ws/5wsb | HTTPS FTP |
-Related structure data
| Related structure data |  5wrpSC  5ws8C  5ws9C  5wsaC  5wscC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: _ / Beg auth comp-ID: THR / Beg label comp-ID: THR / End auth comp-ID: VAL / End label comp-ID: VAL / Refine code: _ / Auth seq-ID: 2 - 472 / Label seq-ID: 5 - 475
NCS ensembles :
|
- Components
Components
-Protein / Sugars , 2 types, 8 molecules ABCD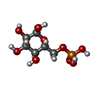

| #1: Protein | Mass: 51014.336 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria)Strain: ATCC 25618 / H37Rv / Gene: pyk, pykA, Rv1617, MTCY01B2.09 / Plasmid: pYUB28b-TEV / Production host:  #3: Sugar | ChemComp-G6P / |
|---|
-Non-polymers , 6 types, 425 molecules 

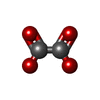








| #2: Chemical | ChemComp-AMP / #4: Chemical | ChemComp-MG / #5: Chemical | ChemComp-OXL / #6: Chemical | ChemComp-K / | #7: Chemical | #8: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.23 Å3/Da / Density % sol: 61.88 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 7.2 Details: 13% PEG 8000, 20% glycerol, 50 mM triethanolamine-HCl (TEA) buffer pH 7.2, 100 mM KCl, 50 mM MgCl2, 5 mM oxalate, 5 mM ATP, 5 mM AMP, 5 mM D-glucose 6-phosphate |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Australian Synchrotron Australian Synchrotron  / Beamline: MX1 / Wavelength: 0.9537 Å / Beamline: MX1 / Wavelength: 0.9537 Å | |||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 210r / Detector: CCD / Date: Apr 28, 2016 | |||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.9537 Å / Relative weight: 1 | |||||||||||||||||||||||||
| Reflection twin |
| |||||||||||||||||||||||||
| Reflection | Resolution: 2.25→62.82 Å / Num. obs: 120301 / % possible obs: 99.3 % / Redundancy: 5.1 % / Net I/σ(I): 9.9 | |||||||||||||||||||||||||
| Reflection shell | Resolution: 2.25→2.29 Å / Redundancy: 2.8 % / Mean I/σ(I) obs: 1.4 / % possible all: 92.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5WRP Resolution: 2.25→62.82 Å / Cor.coef. Fo:Fc: 0.965 / Cor.coef. Fo:Fc free: 0.947 / SU B: 5.799 / SU ML: 0.082 / Cross valid method: THROUGHOUT / ESU R: 0.047 / ESU R Free: 0.04 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 36.436 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 2.25→62.82 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj










