[English] 日本語
 Yorodumi
Yorodumi- PDB-5vvn: Structure of bovine endothelial nitric oxide synthase heme domain... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5vvn | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 4-(2-(((2-Aminoquinolin-7-yl)methyl)amino)ethyl)benzonitrile | ||||||
 Components Components | Nitric oxide synthase, endothelial | ||||||
 Keywords Keywords | OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR / nitric oxide synthase / inhibitor complex / heme enzyme / OXIDOREDUCTASE-OXIDOREDUCTASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of relaxation of smooth muscle / cellular response to laminar fluid shear stress / negative regulation of leukocyte cell-cell adhesion / nitric oxide mediated signal transduction / nitric-oxide synthase (NADPH) / nitric-oxide synthase activity / L-arginine catabolic process / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / negative regulation of blood pressure / response to hormone ...positive regulation of relaxation of smooth muscle / cellular response to laminar fluid shear stress / negative regulation of leukocyte cell-cell adhesion / nitric oxide mediated signal transduction / nitric-oxide synthase (NADPH) / nitric-oxide synthase activity / L-arginine catabolic process / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / negative regulation of blood pressure / response to hormone / nitric oxide biosynthetic process / mitochondrion organization / caveola / response to peptide hormone / blood coagulation / FMN binding / NADP binding / flavin adenine dinucleotide binding / response to lipopolysaccharide / cytoskeleton / calmodulin binding / heme binding / Golgi apparatus / metal ion binding / nucleus / plasma membrane / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 2.4 Å FOURIER SYNTHESIS / Resolution: 2.4 Å | ||||||
 Authors Authors | Li, H. / Poulos, T.L. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: J. Med. Chem. / Year: 2017 Journal: J. Med. Chem. / Year: 2017Title: Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors. Authors: Pensa, A.V. / Cinelli, M.A. / Li, H. / Chreifi, G. / Mukherjee, P. / Roman, L.J. / Martasek, P. / Poulos, T.L. / Silverman, R.B. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5vvn.cif.gz 5vvn.cif.gz | 345.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5vvn.ent.gz pdb5vvn.ent.gz | 279.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5vvn.json.gz 5vvn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vv/5vvn https://data.pdbj.org/pub/pdb/validation_reports/vv/5vvn ftp://data.pdbj.org/pub/pdb/validation_reports/vv/5vvn ftp://data.pdbj.org/pub/pdb/validation_reports/vv/5vvn | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5vuiC  5vujC  5vukC  5vulC  5vumC  5vunC  5vuoC  5vupC  5vuqC  5vurC  5vusC  5vutC  5vuuC  5vuvC  5vuwC  5vuxC  5vuyC  5vuzC  5vv0C  5vv1C  5vv2C  5vv3C  5vv4C  5vv5C  5vv6C  5vv7C  5vv8C  5vv9C  5vvaC  5vvbC  5vvcC  5vvdC  5vvgC  1nseS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 49727.012 Da / Num. of mol.: 2 / Fragment: UNP residues 40-482 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 7 types, 106 molecules 

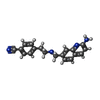










| #2: Chemical | | #3: Chemical | #4: Chemical | #5: Chemical | ChemComp-ACT / #6: Chemical | #7: Chemical | ChemComp-ZN / | #8: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.45 Å3/Da / Density % sol: 49.8 % / Description: cube |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, sitting drop / pH: 6 Details: 20-24% PEG3350, 0.1 M cacodylate, 140-200 mM magnesium acetate, 5 mM TCEP |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL14-1 / Wavelength: 1.087 Å / Beamline: BL14-1 / Wavelength: 1.087 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Apr 5, 2015 / Details: mirrors |
| Radiation | Monochromator: double crystal Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.087 Å / Relative weight: 1 |
| Reflection | Resolution: 2.4→50 Å / Num. obs: 38722 / % possible obs: 99.5 % / Observed criterion σ(I): -3 / Redundancy: 5.7 % / CC1/2: 0.999 / Rmerge(I) obs: 0.076 / Rpim(I) all: 0.052 / Rsym value: 0.076 / Net I/σ(I): 16.1 |
| Reflection shell | Resolution: 2.4→2.54 Å / Redundancy: 5.6 % / Rmerge(I) obs: 1.78 / Mean I/σ(I) obs: 1.1 / Num. unique obs: 3889 / CC1/2: 0.361 / Rpim(I) all: 1.257 / Rsym value: 1.78 / % possible all: 99.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  FOURIER SYNTHESIS FOURIER SYNTHESISStarting model: PDB entry 1NSE Resolution: 2.4→39.232 Å / SU ML: 0.4 / Cross valid method: FREE R-VALUE / σ(F): 0.02 / Phase error: 28.71
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.4→39.232 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj








