+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5vr4 | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA octamer containing 2'-F-4'-OMe U. | ||||||||||||||||||||
 Components Components | RNA (5'-R(* Keywords KeywordsRNA | Function / homology | COBALT TETRAAMMINE ION / RNA |  Function and homology information Function and homology informationBiological species | synthetic construct (others) | Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å MOLECULAR REPLACEMENT / Resolution: 1.5 Å  Authors AuthorsHarp, J.M. / Egli, M. |  Citation Citation Journal: J. Am. Chem. Soc. / Year: 2017 Journal: J. Am. Chem. Soc. / Year: 2017Title: 4'-C-Methoxy-2'-deoxy-2'-fluoro Modified Ribonucleotides Improve Metabolic Stability and Elicit Efficient RNAi-Mediated Gene Silencing. Authors: Malek-Adamian, E. / Guenther, D.C. / Matsuda, S. / Martinez-Montero, S. / Zlatev, I. / Harp, J. / Burai Patrascu, M. / Foster, D.J. / Fakhoury, J. / Perkins, L. / Moitessier, N. / Manoharan, ...Authors: Malek-Adamian, E. / Guenther, D.C. / Matsuda, S. / Martinez-Montero, S. / Zlatev, I. / Harp, J. / Burai Patrascu, M. / Foster, D.J. / Fakhoury, J. / Perkins, L. / Moitessier, N. / Manoharan, R.M. / Taneja, N. / Bisbe, A. / Charisse, K. / Maier, M. / Rajeev, K.G. / Egli, M. / Manoharan, M. / Damha, M.J. History |
|
- Structure visualization
Structure visualization
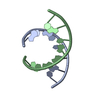
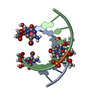
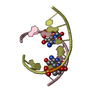
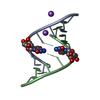
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5vr4.cif.gz 5vr4.cif.gz | 58.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5vr4.ent.gz pdb5vr4.ent.gz | 43.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5vr4.json.gz 5vr4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vr/5vr4 https://data.pdbj.org/pub/pdb/validation_reports/vr/5vr4 ftp://data.pdbj.org/pub/pdb/validation_reports/vr/5vr4 ftp://data.pdbj.org/pub/pdb/validation_reports/vr/5vr4 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3p4aS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||
| 2 | 
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: RNA chain | Mass: 2558.578 Da / Num. of mol.: 4 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) #2: Chemical | ChemComp-CON / | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.08 Å3/Da / Density % sol: 40.51 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 6.6 Details: 0.5 mM RNA, 10 mM Cobalt(III) hexamine, 6 mM sodium chloride, 40 mM potassium chloride, 5% MPD, 20 mM sodium cacodylate equilibrated against reservoir containing 40% MPD Temp details: Room temperature |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 22-ID / Wavelength: 0.97856 Å / Beamline: 22-ID / Wavelength: 0.97856 Å |
| Detector | Type: MARMOSAIC 300 mm CCD / Detector: CCD / Date: Jul 1, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97856 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→30 Å / Num. obs: 26152 / % possible obs: 99.6 % / Redundancy: 7.3 % / Biso Wilson estimate: 25.4 Å2 / Rmerge(I) obs: 0.067 / Rpim(I) all: 0.027 / Net I/σ(I): 21.05 |
| Reflection shell | Resolution: 1.5→1.53 Å / Redundancy: 7.3 % / Rmerge(I) obs: 0.795 / Mean I/σ(I) obs: 2.73 / CC1/2: 0.943 / Rpim(I) all: 0.317 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3P4A Resolution: 1.5→29.481 Å / SU ML: 0.19 / Cross valid method: THROUGHOUT / σ(F): 0.17 / Phase error: 24.16
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.5→29.481 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller






 PDBj
PDBj


