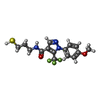
| Entry | Database: PDB / ID: 5vbz
|
|---|
| Title | Crystal Structure of Small Molecule Disulfide 2C07 Bound to H-Ras M72C GppNHp |
|---|
 Components Components | GTPase HRas |
|---|
 Keywords Keywords | HYDROLASE / GTPase / Inhibitor / GDP |
|---|
| Function / homology |  Function and homology information Function and homology information
phospholipase C activator activity / GTPase complex / oncogene-induced cell senescence / positive regulation of miRNA metabolic process / positive regulation of ruffle assembly / T-helper 1 type immune response / positive regulation of wound healing / defense response to protozoan / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / Signaling by RAS GAP mutants ...phospholipase C activator activity / GTPase complex / oncogene-induced cell senescence / positive regulation of miRNA metabolic process / positive regulation of ruffle assembly / T-helper 1 type immune response / positive regulation of wound healing / defense response to protozoan / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / Signaling by RAS GAP mutants / Signaling by RAS GTPase mutants / Activation of RAS in B cells / RAS signaling downstream of NF1 loss-of-function variants / SOS-mediated signalling / Activated NTRK3 signals through RAS / Activated NTRK2 signals through RAS / SHC1 events in ERBB4 signaling / adipose tissue development / positive regulation of protein targeting to membrane / Signalling to RAS / SHC-related events triggered by IGF1R / Activated NTRK2 signals through FRS2 and FRS3 / Schwann cell development / Estrogen-stimulated signaling through PRKCZ / SHC-mediated cascade:FGFR3 / MET activates RAS signaling / SHC-mediated cascade:FGFR2 / SHC-mediated cascade:FGFR4 / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / Signaling by PDGFRA transmembrane, juxtamembrane and kinase domain mutants / Signaling by PDGFRA extracellular domain mutants / Erythropoietin activates RAS / SHC-mediated cascade:FGFR1 / Signaling by FGFR4 in disease / FRS-mediated FGFR3 signaling / Signaling by FLT3 ITD and TKD mutants / FRS-mediated FGFR2 signaling / FRS-mediated FGFR4 signaling / p38MAPK events / FRS-mediated FGFR1 signaling / Signaling by FGFR3 in disease / protein-membrane adaptor activity / Tie2 Signaling / Signaling by FGFR2 in disease / myelination / EPHB-mediated forward signaling / GRB2 events in EGFR signaling / Signaling by FLT3 fusion proteins / SHC1 events in EGFR signaling / FLT3 Signaling / Signaling by FGFR1 in disease / EGFR Transactivation by Gastrin / NCAM signaling for neurite out-growth / CD209 (DC-SIGN) signaling / GRB2 events in ERBB2 signaling / Downstream signal transduction / Insulin receptor signalling cascade / intrinsic apoptotic signaling pathway / SHC1 events in ERBB2 signaling / Ras activation upon Ca2+ influx through NMDA receptor / Constitutive Signaling by Overexpressed ERBB2 / Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants / animal organ morphogenesis / VEGFR2 mediated cell proliferation / positive regulation of epithelial cell proliferation / small monomeric GTPase / regulation of actin cytoskeleton organization / FCERI mediated MAPK activation / Signaling by ERBB2 TMD/JMD mutants / RAF activation / Signaling by SCF-KIT / Constitutive Signaling by EGFRvIII / Signaling by high-kinase activity BRAF mutants / cellular response to gamma radiation / MAP2K and MAPK activation / Signaling by ERBB2 ECD mutants / Signaling by ERBB2 KD Mutants / regulation of long-term neuronal synaptic plasticity / positive regulation of JNK cascade / positive regulation of type II interferon production / positive regulation of fibroblast proliferation / endocytosis / chemotaxis / Regulation of RAS by GAPs / Negative regulation of MAPK pathway / Signaling by RAF1 mutants / RAS processing / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / cellular senescence / GDP binding / Signaling by BRAF and RAF1 fusions / insulin receptor signaling pathway / DAP12 signaling / MAPK cascade / T cell receptor signaling pathway / regulation of cell population proliferation / Constitutive Signaling by Ligand-Responsive EGFR Cancer Variants / RAF/MAP kinase cascadeSimilarity search - Function Small GTPase, Ras-type / Small GTPase Ras domain profile. / Rho (Ras homology) subfamily of Ras-like small GTPases / Ras subfamily of RAS small GTPases / Small GTPase / Ras family / Rab subfamily of small GTPases / Small GTP-binding protein domain / P-loop containing nucleotide triphosphate hydrolases / Rossmann fold ...Small GTPase, Ras-type / Small GTPase Ras domain profile. / Rho (Ras homology) subfamily of Ras-like small GTPases / Ras subfamily of RAS small GTPases / Small GTPase / Ras family / Rab subfamily of small GTPases / Small GTP-binding protein domain / P-loop containing nucleotide triphosphate hydrolases / Rossmann fold / P-loop containing nucleoside triphosphate hydrolase / 3-Layer(aba) Sandwich / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å |
|---|
 Authors Authors | Gentile, D.R. / Jenkins, M.L. / Moss, S.M. / Burke, J.E. / Shokat, K.M. |
|---|
 Citation Citation |  Journal: Cell Chem Biol / Year: 2017 Journal: Cell Chem Biol / Year: 2017
Title: Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Authors: Gentile, D.R. / Rathinaswamy, M.K. / Jenkins, M.L. / Moss, S.M. / Siempelkamp, B.D. / Renslo, A.R. / Burke, J.E. / Shokat, K.M. |
|---|
| History | | Deposition | Mar 30, 2017 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Oct 25, 2017 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Nov 1, 2017 | Group: Database references / Category: citation / citation_author
Item: _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title |
|---|
| Revision 1.2 | Jan 3, 2018 | Group: Database references / Category: citation
Item: _citation.journal_volume / _citation.page_first / _citation.page_last |
|---|
| Revision 1.3 | Oct 4, 2023 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_conn / struct_conn_type
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn_type.id |
|---|
| Revision 1.4 | Nov 20, 2024 | Group: Structure summary / Category: pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å
MOLECULAR REPLACEMENT / Resolution: 2.2 Å  Authors
Authors Citation
Citation Journal: Cell Chem Biol / Year: 2017
Journal: Cell Chem Biol / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5vbz.cif.gz
5vbz.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5vbz.ent.gz
pdb5vbz.ent.gz PDB format
PDB format 5vbz.json.gz
5vbz.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/vb/5vbz
https://data.pdbj.org/pub/pdb/validation_reports/vb/5vbz ftp://data.pdbj.org/pub/pdb/validation_reports/vb/5vbz
ftp://data.pdbj.org/pub/pdb/validation_reports/vb/5vbz

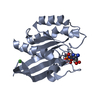
 Links
Links Assembly
Assembly



 Components
Components Homo sapiens (human) / Gene: HRAS, HRAS1 / Production host:
Homo sapiens (human) / Gene: HRAS, HRAS1 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ALS
ALS  / Beamline: 8.2.2 / Wavelength: 1 Å
/ Beamline: 8.2.2 / Wavelength: 1 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller










 PDBj
PDBj