[English] 日本語
 Yorodumi
Yorodumi- PDB-5sys: c-Src V281C bound to N-[3-({6-[(1E)-2-cyano-3-(methylamino)-3-oxo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5sys | ||||||
|---|---|---|---|---|---|---|---|
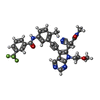
| Title | c-Src V281C bound to N-[3-({6-[(1E)-2-cyano-3-(methylamino)-3-oxoprop-1-en-1-yl]-7-(2-methoxyethyl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl}ethynyl)-4-methylphenyl]-3-(trifluoromethyl)benzamide inhibitor | ||||||
 Components Components | Proto-oncogene tyrosine-protein kinase Src | ||||||
 Keywords Keywords | TRANSFERASE/TRANSFERASE INHIBITOR / DFG-out / covalently-linked inihibitor / TRANSFERASE-TRANSFERASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationSignaling by ERBB2 / Nuclear signaling by ERBB4 / Signaling by SCF-KIT / Regulation of KIT signaling / Signaling by EGFR / GAB1 signalosome / Regulation of gap junction activity / FCGR activation / PECAM1 interactions / Co-stimulation by CD28 ...Signaling by ERBB2 / Nuclear signaling by ERBB4 / Signaling by SCF-KIT / Regulation of KIT signaling / Signaling by EGFR / GAB1 signalosome / Regulation of gap junction activity / FCGR activation / PECAM1 interactions / Co-stimulation by CD28 / Co-inhibition by CTLA4 / EPHA-mediated growth cone collapse / Ephrin signaling / G alpha (i) signalling events / GP1b-IX-V activation signalling / Thrombin signalling through proteinase activated receptors (PARs) / VEGFR2 mediated cell proliferation / RET signaling / Receptor Mediated Mitophagy / ADP signalling through P2Y purinoceptor 1 / RAF activation / PIP3 activates AKT signaling / EPH-ephrin mediated repulsion of cells / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / : / Downstream signal transduction / Downregulation of ERBB4 signaling / Cyclin D associated events in G1 / Regulation of RUNX3 expression and activity / Degradation of CDH1 / MAP2K and MAPK activation / Integrin signaling / GRB2:SOS provides linkage to MAPK signaling for Integrins / : / MET activates PTK2 signaling / Extra-nuclear estrogen signaling / EPHB-mediated forward signaling / p130Cas linkage to MAPK signaling for integrins / VEGFA-VEGFR2 Pathway / connexin binding / negative regulation of intrinsic apoptotic signaling pathway / progesterone receptor signaling pathway / immune system process / negative regulation of extrinsic apoptotic signaling pathway / non-membrane spanning protein tyrosine kinase activity / non-specific protein-tyrosine kinase / epidermal growth factor receptor signaling pathway / cell-cell junction / cell junction / protein tyrosine kinase activity / protein phosphatase binding / cytoskeleton / cell differentiation / cell adhesion / endosome membrane / regulation of cell cycle / mitochondrial inner membrane / signaling receptor binding / focal adhesion / heme binding / perinuclear region of cytoplasm / protein-containing complex / ATP binding / membrane / nucleus / plasma membrane / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.8 Å MOLECULAR REPLACEMENT / Resolution: 2.8 Å | ||||||
 Authors Authors | Dieter, E.M. / Merritt, E.A. / Maly, D.J. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Mol.Cell Journal: Mol.CellTitle: A combined approach reveals a regulatory mechanism coupling Src's kinase activity, localization, and phosphotransferase-independent functions Authors: Register, A.C. / Maly, D.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5sys.cif.gz 5sys.cif.gz | 121.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5sys.ent.gz pdb5sys.ent.gz | 93.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5sys.json.gz 5sys.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sy/5sys https://data.pdbj.org/pub/pdb/validation_reports/sy/5sys ftp://data.pdbj.org/pub/pdb/validation_reports/sy/5sys ftp://data.pdbj.org/pub/pdb/validation_reports/sy/5sys | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5swhC  5tehC  3uqgS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||
| 2 | 
| ||||||||||||||||||
| Unit cell |
| ||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: _ / Ens-ID: 1 / Beg auth comp-ID: ASP / Beg label comp-ID: ASP / End auth comp-ID: LEU / End label comp-ID: LEU / Refine code: _ / Auth seq-ID: 258 - 533 / Label seq-ID: 11 - 286
|
- Components
Components
| #1: Protein | Mass: 32730.656 Da / Num. of mol.: 2 / Mutation: V281C Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: P00523, non-specific protein-tyrosine kinase #2: Chemical | #3: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.02 Å3/Da / Density % sol: 59.21 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop Details: 100 mM MES pH 6, 8% PEG 3350, 10 mM NaOAc, 14% glycerol, 10 mM DTT |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-2 / Wavelength: 0.97946 Å / Beamline: BL9-2 / Wavelength: 0.97946 Å | ||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Jun 2, 2016 | ||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.97946 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.8→73.94 Å / Num. obs: 16285 / % possible obs: 86.6 % / Redundancy: 3.3 % / CC1/2: 0.987 / Rmerge(I) obs: 0.176 / Rpim(I) all: 0.111 / Rrim(I) all: 0.209 / Net I/σ(I): 5.7 / Num. measured all: 54004 / Scaling rejects: 27 | ||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / Rejects: _
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3uqg Resolution: 2.8→73.94 Å / Cor.coef. Fo:Fc: 0.819 / Cor.coef. Fo:Fc free: 0.792 / SU B: 32.468 / SU ML: 0.544 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R Free: 0.527 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : REFINED INDIVIDUALLY
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 218.95 Å2 / Biso mean: 64.488 Å2 / Biso min: 2 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.8→73.94 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Ens-ID: 1 / Number: 16560 / Refine-ID: X-RAY DIFFRACTION / Type: interatomic distance / Rms dev position: 0.05 Å / Weight position: 0.05
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.801→2.874 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller















 PDBj
PDBj