| 登録情報 | データベース: PDB / ID: 5svh
|
|---|
| タイトル | Crystal structure of the KIX domain of CBP in complex with a MLL/c-Myb chimera |
|---|
 要素 要素 | - CREB-binding protein
- MLL/c-Myb chimera
|
|---|
 キーワード キーワード | TRANSCRIPTION / CBP / complex |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
negative regulation of DNA methylation-dependent heterochromatin formation / protein-cysteine methyltransferase activity / [histone H3]-lysine4 N-methyltransferase / histone H3K4 monomethyltransferase activity / response to potassium ion / unmethylated CpG binding / histone H3K4 trimethyltransferase activity / peptide lactyltransferase (CoA-dependent) activity / NFE2L2 regulating ER-stress associated genes / Activation of the TFAP2 (AP-2) family of transcription factors ...negative regulation of DNA methylation-dependent heterochromatin formation / protein-cysteine methyltransferase activity / [histone H3]-lysine4 N-methyltransferase / histone H3K4 monomethyltransferase activity / response to potassium ion / unmethylated CpG binding / histone H3K4 trimethyltransferase activity / peptide lactyltransferase (CoA-dependent) activity / NFE2L2 regulating ER-stress associated genes / Activation of the TFAP2 (AP-2) family of transcription factors / NFE2L2 regulating inflammation associated genes / histone H3K18 acetyltransferase activity / N-terminal peptidyl-lysine acetylation / histone H3K27 acetyltransferase activity / LRR FLII-interacting protein 1 (LRRFIP1) activates type I IFN production / NFE2L2 regulates pentose phosphate pathway genes / regulation of smoothened signaling pathway / definitive hemopoiesis / regulation of short-term neuronal synaptic plasticity / NFE2L2 regulating MDR associated enzymes / MRF binding / histone H3K4 methyltransferase activity / RUNX1 regulates transcription of genes involved in differentiation of myeloid cells / Regulation of gene expression in late stage (branching morphogenesis) pancreatic bud precursor cells / Regulation of FOXO transcriptional activity by acetylation / RUNX3 regulates NOTCH signaling / NOTCH4 Intracellular Domain Regulates Transcription / Nuclear events mediated by NFE2L2 / Regulation of NFE2L2 gene expression / Regulation of gene expression by Hypoxia-inducible Factor / anterior/posterior pattern specification / embryonic hemopoiesis / T-helper 2 cell differentiation / negative regulation of transcription by RNA polymerase I / NOTCH3 Intracellular Domain Regulates Transcription / TRAF6 mediated IRF7 activation / NFE2L2 regulating tumorigenic genes / NFE2L2 regulating anti-oxidant/detoxification enzymes / embryonic digit morphogenesis / protein-lysine-acetyltransferase activity / protein acetylation / homeostatic process / Notch-HLH transcription pathway / Formation of WDR5-containing histone-modifying complexes / Formation of paraxial mesoderm / positive regulation of transforming growth factor beta receptor signaling pathway / acetyltransferase activity / FOXO-mediated transcription of cell death genes / stimulatory C-type lectin receptor signaling pathway / Zygotic genome activation (ZGA) / exploration behavior / TP53 Regulates Transcription of Genes Involved in Cytochrome C Release / histone methyltransferase complex / minor groove of adenine-thymine-rich DNA binding / MLL1 complex / histone acetyltransferase complex / membrane depolarization / canonical NF-kappaB signal transduction / Attenuation phase / cellular response to transforming growth factor beta stimulus / negative regulation of fibroblast proliferation / histone acetyltransferase activity / cellular response to nutrient levels / histone acetyltransferase / positive regulation of double-strand break repair via homologous recombination / regulation of cellular response to heat / spleen development / : / NPAS4 regulates expression of target genes / Regulation of lipid metabolism by PPARalpha / 転移酵素; アシル基を移すもの; アミノアシル基以外のアシル基を移すもの / Transcriptional and post-translational regulation of MITF-M expression and activity / CD209 (DC-SIGN) signaling / homeostasis of number of cells within a tissue / BMAL1:CLOCK,NPAS2 activates circadian expression / transcription initiation-coupled chromatin remodeling / SUMOylation of transcription cofactors / Activation of gene expression by SREBF (SREBP) / 転移酵素; 一炭素原子の基を移すもの; メチル基を移すもの / post-embryonic development / circadian regulation of gene expression / Heme signaling / Transcriptional activation of mitochondrial biogenesis / Formation of the beta-catenin:TCF transactivating complex / PPARA activates gene expression / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / Cytoprotection by HMOX1 / protein destabilization / visual learning / protein modification process / chromatin DNA binding / Evasion by RSV of host interferon responses / NOTCH1 Intracellular Domain Regulates Transcription / Transcriptional regulation of white adipocyte differentiation / PKMTs methylate histone lysines / positive regulation of protein localization to nucleus / Pre-NOTCH Transcription and Translation / Constitutive Signaling by NOTCH1 PEST Domain Mutants / Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants / transcription coactivator binding類似検索 - 分子機能 Coactivator CBP, KIX domain / Transcription regulator Wos2-domain / LMSTEN motif / C-myb, C-terminal / C-myb, C-terminal / : / KMT2A, ePHD domain / KMT2A, PHD domain 1 / KMT2A, PHD domain 2 / KMT2A, PHD domain 3 ...Coactivator CBP, KIX domain / Transcription regulator Wos2-domain / LMSTEN motif / C-myb, C-terminal / C-myb, C-terminal / : / KMT2A, ePHD domain / KMT2A, PHD domain 1 / KMT2A, PHD domain 2 / KMT2A, PHD domain 3 / Methyltransferase, trithorax / : / FY-rich, N-terminal / F/Y-rich N-terminus / PHD-like zinc-binding domain / FYR domain FYRN motif profile. / "FY-rich" domain, N-terminal region / FY-rich, C-terminal / F/Y rich C-terminus / FYR domain FYRC motif profile. / "FY-rich" domain, C-terminal region / CXXC zinc finger domain / Zinc finger, CXXC-type / Zinc finger CXXC-type profile. / Cysteine-rich motif following a subset of SET domains / Nuclear receptor coactivator, CREB-bp-like, interlocking / Nuclear receptor coactivator, CREB-bp-like, interlocking domain superfamily / Creb binding / Zinc finger, TAZ-type / TAZ domain superfamily / TAZ zinc finger / Zinc finger TAZ-type profile. / TAZ zinc finger, present in p300 and CBP / : / Histone acetyltransferase p300-like, PHD domain / Coactivator CBP, KIX domain / CREB-binding protein/p300, atypical RING domain / CBP/p300-type histone acetyltransferase domain / CBP/p300, atypical RING domain superfamily / KIX domain / CREB-binding protein/p300, atypical RING domain / KIX domain profile. / CBP/p300-type histone acetyltransferase (HAT) domain profile. / Histone acetyltransferase Rtt109/CBP / Histone acetylation protein / Histone acetylation protein / Coactivator CBP, KIX domain superfamily / Post-SET domain / Post-SET domain profile. / Myb-type HTH DNA-binding domain profile. / Serum Albumin; Chain A, Domain 1 / Zinc finger ZZ-type signature. / Myb domain / Zinc-binding domain, present in Dystrophin, CREB-binding protein. / Zinc finger, ZZ type / Zinc finger, ZZ-type / Zinc finger, ZZ-type superfamily / Zinc finger ZZ-type profile. / Myb-like DNA-binding domain / Extended PHD (ePHD) domain / Extended PHD (ePHD) domain profile. / SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain / SET domain / SET domain profile. / SET domain superfamily / SET domain / SANT SWI3, ADA2, N-CoR and TFIIIB'' DNA-binding domains / SANT/Myb domain / Nuclear receptor coactivator, interlocking / PHD-finger / Zinc finger PHD-type signature. / Zinc finger PHD-type profile. / Zinc finger, PHD-finger / Zinc finger, PHD-type / PHD zinc finger / Zinc finger, FYVE/PHD-type / Homeobox-like domain superfamily / Bromodomain, conserved site / Bromodomain signature. / Bromodomain / bromo domain / Bromodomain / Bromodomain (BrD) profile. / Bromodomain-like superfamily / Zinc finger, RING/FYVE/PHD-type / Orthogonal Bundle / Mainly Alpha類似検索 - ドメイン・相同性 Transcriptional activator Myb / Histone-lysine N-methyltransferase 2A / CREB-binding protein類似検索 - 構成要素 |
|---|
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト)
  Gallus gallus (ニワトリ) Gallus gallus (ニワトリ) |
|---|
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 2.05 Å 分子置換 / 解像度: 2.05 Å |
|---|
 データ登録者 データ登録者 | Langelaan, D.N. / Smith, S.P. / Allingham, J.A. |
|---|
| 資金援助 |  カナダ, 1件 カナダ, 1件 | 組織 | 認可番号 | 国 |
|---|
| Canadian Institutes of Health Research (CIHR) | |  カナダ カナダ |
|
|---|
 引用 引用 |  ジャーナル: To Be Published ジャーナル: To Be Published
タイトル: Design of a nanomolar affinity ligand to the KIX domain of CBP
著者: Langelaan, D.N. / Smith, S.P. |
|---|
| 履歴 | | 登録 | 2016年8月6日 | 登録サイト: RCSB / 処理サイト: RCSB |
|---|
| 改定 1.0 | 2018年5月9日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2018年10月17日 | Group: Data collection / Source and taxonomy / Structure summary
カテゴリ: entity / entity_src_gen / Item: _entity.pdbx_fragment |
|---|
| 改定 1.2 | 2020年1月8日 | Group: Author supporting evidence / カテゴリ: pdbx_audit_support / Item: _pdbx_audit_support.funding_organization |
|---|
| 改定 1.3 | 2023年10月4日 | Group: Data collection / Database references / Refinement description
カテゴリ: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト)
 X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 2.05 Å
分子置換 / 解像度: 2.05 Å  データ登録者
データ登録者 カナダ, 1件
カナダ, 1件  引用
引用 ジャーナル: To Be Published
ジャーナル: To Be Published 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 5svh.cif.gz
5svh.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb5svh.ent.gz
pdb5svh.ent.gz PDB形式
PDB形式 5svh.json.gz
5svh.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 5svh_validation.pdf.gz
5svh_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 5svh_full_validation.pdf.gz
5svh_full_validation.pdf.gz 5svh_validation.xml.gz
5svh_validation.xml.gz 5svh_validation.cif.gz
5svh_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/sv/5svh
https://data.pdbj.org/pub/pdb/validation_reports/sv/5svh ftp://data.pdbj.org/pub/pdb/validation_reports/sv/5svh
ftp://data.pdbj.org/pub/pdb/validation_reports/sv/5svh
 リンク
リンク 集合体
集合体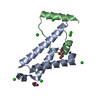


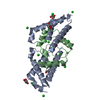
 要素
要素 Homo sapiens (ヒト) / 遺伝子: CREBBP, CBP / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: CREBBP, CBP / 発現宿主: 
 Homo sapiens (ヒト), (組換発現)
Homo sapiens (ヒト), (組換発現) 

 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  APS
APS  / ビームライン: 23-ID-B / 波長: 1.0332 Å
/ ビームライン: 23-ID-B / 波長: 1.0332 Å 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー











 PDBj
PDBj





















