| Entry | Database: PDB / ID: 5or1
|
|---|
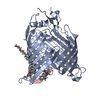
| Title | BamA structure of Salmonella enterica |
|---|
 Components Components | Outer membrane protein assembly factor BamA |
|---|
 Keywords Keywords | MEMBRANE PROTEIN / Outer membrane protein |
|---|
| Function / homology |  Function and homology information Function and homology information
Bam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / protein insertion into membraneSimilarity search - Function membrane protein fhac: a member of the omp85/tpsb transporter family / Outer membrane protein assembly factor BamA / POTRA domain, BamA/TamA-like / Surface antigen variable number repeat / Surface antigen D15-like / POTRA domain / POTRA domain profile. / Bacterial surface antigen (D15) / Omp85 superfamily domain / Porin ...membrane protein fhac: a member of the omp85/tpsb transporter family / Outer membrane protein assembly factor BamA / POTRA domain, BamA/TamA-like / Surface antigen variable number repeat / Surface antigen D15-like / POTRA domain / POTRA domain profile. / Bacterial surface antigen (D15) / Omp85 superfamily domain / Porin / Beta Barrel / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Salmonella typhimurium (bacteria) Salmonella typhimurium (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.92 Å MOLECULAR REPLACEMENT / Resolution: 2.92 Å |
|---|
 Authors Authors | Dong, C. / Gu, Y. |
|---|
 Citation Citation |  Journal: Biochem. J. / Year: 2017 Journal: Biochem. J. / Year: 2017
Title: BamA beta 16C strand and periplasmic turns are critical for outer membrane protein insertion and assembly.
Authors: Gu, Y. / Zeng, Y. / Wang, Z. / Dong, C. |
|---|
| History | | Deposition | Aug 14, 2017 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Feb 14, 2018 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Oct 16, 2019 | Group: Data collection / Category: reflns_shell |
|---|
| Revision 1.2 | May 8, 2024 | Group: Data collection / Database references / Category: chem_comp_atom / chem_comp_bond / database_2
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Salmonella typhimurium (bacteria)
Salmonella typhimurium (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.92 Å
MOLECULAR REPLACEMENT / Resolution: 2.92 Å  Authors
Authors Citation
Citation Journal: Biochem. J. / Year: 2017
Journal: Biochem. J. / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5or1.cif.gz
5or1.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5or1.ent.gz
pdb5or1.ent.gz PDB format
PDB format 5or1.json.gz
5or1.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/or/5or1
https://data.pdbj.org/pub/pdb/validation_reports/or/5or1 ftp://data.pdbj.org/pub/pdb/validation_reports/or/5or1
ftp://data.pdbj.org/pub/pdb/validation_reports/or/5or1 Links
Links Assembly
Assembly
 Components
Components Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)
Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I04-1 / Wavelength: 0.92 Å
/ Beamline: I04-1 / Wavelength: 0.92 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 2.92→107.42 Å / Cor.coef. Fo:Fc: 0.855 / Cor.coef. Fo:Fc free: 0.874 / SU B: 20.586 / SU ML: 0.397 / Cross valid method: THROUGHOUT / ESU R: 1.524 / ESU R Free: 0.494 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
MOLECULAR REPLACEMENT / Resolution: 2.92→107.42 Å / Cor.coef. Fo:Fc: 0.855 / Cor.coef. Fo:Fc free: 0.874 / SU B: 20.586 / SU ML: 0.397 / Cross valid method: THROUGHOUT / ESU R: 1.524 / ESU R Free: 0.494 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS Movie
Movie Controller
Controller












 PDBj
PDBj