[English] 日本語
 Yorodumi
Yorodumi- PDB-5oce: THE MOLECULAR MECHANISM OF SUBSTRATE RECOGNITION AND CATALYSIS OF... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5oce | ||||||
|---|---|---|---|---|---|---|---|
| Title | THE MOLECULAR MECHANISM OF SUBSTRATE RECOGNITION AND CATALYSIS OF THE MEMBRANE ACYLTRANSFERASE PatA -- Complex of PatA with palmitate, mannose, and palmitoyl-6-mannose | ||||||
 Components Components | Phosphatidylinositol mannoside acyltransferase | ||||||
 Keywords Keywords | TRANSFERASE / acyltransferase / glycolipid biosynthesis | ||||||
| Function / homology |  Function and homology information Function and homology informationphosphatidylinositol dimannoside acyltransferase / glycolipid biosynthetic process / phosphatidylinositol metabolic process / phospholipid biosynthetic process / acyltransferase activity / plasma membrane Similarity search - Function | ||||||
| Biological species |  Mycobacterium smegmatis (bacteria) Mycobacterium smegmatis (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.41 Å MOLECULAR REPLACEMENT / Resolution: 2.41 Å | ||||||
 Authors Authors | Albesa-Jove, D. / Tersa, M. / Guerin, M.E. | ||||||
 Citation Citation |  Journal: ACS Chem. Biol. / Year: 2018 Journal: ACS Chem. Biol. / Year: 2018Title: The Molecular Mechanism of Substrate Recognition and Catalysis of the Membrane Acyltransferase PatA from Mycobacteria. Authors: Tersa, M. / Raich, L. / Albesa-Jove, D. / Trastoy, B. / Prandi, J. / Gilleron, M. / Rovira, C. / Guerin, M.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5oce.cif.gz 5oce.cif.gz | 355.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5oce.ent.gz pdb5oce.ent.gz | 294.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5oce.json.gz 5oce.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/oc/5oce https://data.pdbj.org/pub/pdb/validation_reports/oc/5oce ftp://data.pdbj.org/pub/pdb/validation_reports/oc/5oce ftp://data.pdbj.org/pub/pdb/validation_reports/oc/5oce | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5f2tS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
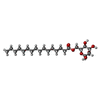
| #1: Protein | Mass: 33591.062 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Gene: MSMEG_2934, MSMEI_2860 / Production host:  Mycobacterium smegmatis (bacteria) Mycobacterium smegmatis (bacteria)References: UniProt: A0QWG5, Transferases; Acyltransferases; Transferring groups other than aminoacyl groups #2: Chemical | ChemComp-PLM / | #3: Sugar | ChemComp-BMA / | #4: Chemical | #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.03 Å3/Da / Density % sol: 39.33 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop Details: The crystal of PatA was obtained by mixing 0.25ul of the protein (5 mg ml-1) in 5 mM D-(+)-mannose and 20 mM Tris-HCl buffer pH 7.5 with 0.25ul of a mother liquor containing, 100 mM Tris-HCl ...Details: The crystal of PatA was obtained by mixing 0.25ul of the protein (5 mg ml-1) in 5 mM D-(+)-mannose and 20 mM Tris-HCl buffer pH 7.5 with 0.25ul of a mother liquor containing, 100 mM Tris-HCl pH 7.0, 225 mM MgCl2 and 16% (w/v) PEG 8,000. PH range: 7.0-7.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I24 / Wavelength: 0.9686 Å / Beamline: I24 / Wavelength: 0.9686 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: May 21, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9686 Å / Relative weight: 1 |
| Reflection | Resolution: 2.41→28.77 Å / Num. obs: 40218 / % possible obs: 97.17 % / Redundancy: 3.3 % / CC1/2: 0.989 / Rmerge(I) obs: 0.1125 / Net I/σ(I): 12.76 |
| Reflection shell | Resolution: 2.41→2.5 Å / Redundancy: 3.3 % / Rmerge(I) obs: 0.4939 / Num. unique obs: 3754 / CC1/2: 0.773 / % possible all: 90.55 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5F2T Resolution: 2.41→28.77 Å / Cross valid method: FREE R-VALUE / σ(F): 1.45 / Phase error: 22.02
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 44.52 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.41→28.77 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj