[English] 日本語
 Yorodumi
Yorodumi- PDB-5mzp: Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5mzp | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with caffeine at 2.1A resolution | ||||||
 Components Components | Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / G-PROTEIN-COUPLED RECEPTOR / INTEGRAL MEMBRANE PROTEIN / CHIMERA / THERMOSTABILIZING MUTATIONS | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of norepinephrine secretion / positive regulation of acetylcholine secretion, neurotransmission / negative regulation of alpha-beta T cell activation / positive regulation of circadian sleep/wake cycle, sleep / Adenosine P1 receptors / G protein-coupled adenosine receptor activity / response to purine-containing compound / G protein-coupled adenosine receptor signaling pathway / NGF-independant TRKA activation / Surfactant metabolism ...regulation of norepinephrine secretion / positive regulation of acetylcholine secretion, neurotransmission / negative regulation of alpha-beta T cell activation / positive regulation of circadian sleep/wake cycle, sleep / Adenosine P1 receptors / G protein-coupled adenosine receptor activity / response to purine-containing compound / G protein-coupled adenosine receptor signaling pathway / NGF-independant TRKA activation / Surfactant metabolism / sensory perception / positive regulation of urine volume / synaptic transmission, dopaminergic / type 5 metabotropic glutamate receptor binding / negative regulation of vascular permeability / synaptic transmission, cholinergic / positive regulation of glutamate secretion / intermediate filament / presynaptic active zone / response to caffeine / blood circulation / eating behavior / inhibitory postsynaptic potential / alpha-actinin binding / regulation of calcium ion transport / asymmetric synapse / axolemma / membrane depolarization / phagocytosis / cellular defense response / prepulse inhibition / positive regulation of synaptic transmission, glutamatergic / neuron projection morphogenesis / astrocyte activation / presynaptic modulation of chemical synaptic transmission / response to amphetamine / central nervous system development / positive regulation of long-term synaptic potentiation / positive regulation of synaptic transmission, GABAergic / positive regulation of protein secretion / positive regulation of apoptotic signaling pathway / regulation of mitochondrial membrane potential / synaptic transmission, glutamatergic / excitatory postsynaptic potential / apoptotic signaling pathway / locomotory behavior / electron transport chain / negative regulation of inflammatory response / vasodilation / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / blood coagulation / cell-cell signaling / presynaptic membrane / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / negative regulation of neuron apoptotic process / postsynaptic membrane / calmodulin binding / electron transfer activity / periplasmic space / positive regulation of ERK1 and ERK2 cascade / iron ion binding / response to xenobiotic stimulus / inflammatory response / negative regulation of cell population proliferation / neuronal cell body / heme binding / apoptotic process / dendrite / regulation of DNA-templated transcription / lipid binding / protein-containing complex binding / glutamatergic synapse / enzyme binding / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | ||||||
 Authors Authors | Cheng, K.Y.R. / Segala, E. / Robertson, N. / Deflorian, F. / Dore, A.S. / Errey, J.C. / Fiez-Vandal, C. / Marshall, F.H. / Cooke, R.M. | ||||||
 Citation Citation |  Journal: Structure / Year: 2017 Journal: Structure / Year: 2017Title: Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity. Authors: Cheng, R.K.Y. / Segala, E. / Robertson, N. / Deflorian, F. / Dore, A.S. / Errey, J.C. / Fiez-Vandal, C. / Marshall, F.H. / Cooke, R.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5mzp.cif.gz 5mzp.cif.gz | 189 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5mzp.ent.gz pdb5mzp.ent.gz | 147.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5mzp.json.gz 5mzp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mz/5mzp https://data.pdbj.org/pub/pdb/validation_reports/mz/5mzp ftp://data.pdbj.org/pub/pdb/validation_reports/mz/5mzp ftp://data.pdbj.org/pub/pdb/validation_reports/mz/5mzp | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5mzjC  5n2rC  5n2sC  5iu4S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 47996.746 Da / Num. of mol.: 1 Mutation: A54L, T88A, R107A, K122A, N154A, L202A, M1007W, L235A, V239A, S277A, G318A,A54L, T88A, R107A, K122A, N154A, L202A, M1007W, L235A, V239A, S277A, G318A,A54L, T88A, R107A, K122A, N154A, L202A, ...Mutation: A54L, T88A, R107A, K122A, N154A, L202A, M1007W, L235A, V239A, S277A, G318A,A54L, T88A, R107A, K122A, N154A, L202A, M1007W, L235A, V239A, S277A, G318A,A54L, T88A, R107A, K122A, N154A, L202A, M1007W, L235A, V239A, S277A, G318A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human), (gene. exp.) Homo sapiens (human), (gene. exp.)  Gene: ADORA2A, ADORA2, cybC / Plasmid: PFAST-BAC / Production host:  Trichoplusia ni (cabbage looper) / Variant (production host): Tni PRO / References: UniProt: P29274, UniProt: P0ABE7 Trichoplusia ni (cabbage looper) / Variant (production host): Tni PRO / References: UniProt: P29274, UniProt: P0ABE7 |
|---|
-Non-polymers , 7 types, 209 molecules 











| #2: Chemical | ChemComp-NA / | ||||||||
|---|---|---|---|---|---|---|---|---|---|
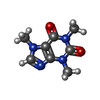
| #3: Chemical | ChemComp-CFF / | ||||||||
| #4: Chemical | ChemComp-CLR / #5: Chemical | ChemComp-OLA / #6: Chemical | ChemComp-OLC / ( #7: Chemical | ChemComp-TAR / | #8: Water | ChemComp-HOH / | |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.58 Å3/Da / Density % sol: 52.36 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: lipidic cubic phase / pH: 5.5 Details: 0.1M MES PH 5.5, 0.2M K/NA TARTRATE, 27.5-40% PEG400, 0.5-1% (V/V) (+/-)-2-METHYL-2,4-PENTANEDIOL |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I24 / Wavelength: 0.96862 Å / Beamline: I24 / Wavelength: 0.96862 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Mar 25, 2016 / Details: (DOUBLE) KB MIRROR PAIR |
| Radiation | Monochromator: DOUBLE SI (111) CRYSTAL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.96862 Å / Relative weight: 1 |
| Reflection | Resolution: 2.1→89.94 Å / Num. obs: 29678 / % possible obs: 98.7 % / Redundancy: 4.8 % / CC1/2: 0.996 / Rmerge(I) obs: 0.13 / Net I/σ(I): 8.2 |
| Reflection shell | Resolution: 2.1→2.16 Å / Redundancy: 4.9 % / Rmerge(I) obs: 0.977 / Mean I/σ(I) obs: 1.4 / Num. unique obs: 2379 / CC1/2: 0.454 / Rpim(I) all: 0.532 / % possible all: 99.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5IU4 Resolution: 2.1→38.527 Å / SU ML: 0.21 / Cross valid method: FREE R-VALUE / σ(F): 1.36 / Phase error: 21.33
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→38.527 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | S33: -0 Å ° / Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj























