[English] 日本語
 Yorodumi
Yorodumi- PDB-5kmw: TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5kmw | ||||||
|---|---|---|---|---|---|---|---|
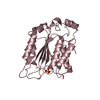
| Title | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex | ||||||
 Components Components | Beta-lactamase Toho-1 | ||||||
 Keywords Keywords | HYDROLASE / Class A beta-lactamase / substrate recognition / acyl-enzyme | ||||||
| Function / homology |  Function and homology information Function and homology informationbeta-lactam antibiotic catabolic process / beta-lactamase activity / beta-lactamase / response to antibiotic Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.1 Å MOLECULAR REPLACEMENT / Resolution: 1.1 Å | ||||||
 Authors Authors | Coates, L. / Langan, P.S. / Vandavasi, V.G. / Weiss, K.L. / Cooper, J.B. / Ginell, S.L. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: to be published Journal: to be publishedTitle: TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex Authors: Coates, L. / Langan, P.S. / Vandavasi, V.G. / Weiss, K.L. / Cooper, J.B. / Ginell, S.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5kmw.cif.gz 5kmw.cif.gz | 140.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5kmw.ent.gz pdb5kmw.ent.gz | 107.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5kmw.json.gz 5kmw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  5kmw_validation.pdf.gz 5kmw_validation.pdf.gz | 1.4 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  5kmw_full_validation.pdf.gz 5kmw_full_validation.pdf.gz | 1.4 MB | Display | |
| Data in XML |  5kmw_validation.xml.gz 5kmw_validation.xml.gz | 19.5 KB | Display | |
| Data in CIF |  5kmw_validation.cif.gz 5kmw_validation.cif.gz | 30.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/km/5kmw https://data.pdbj.org/pub/pdb/validation_reports/km/5kmw ftp://data.pdbj.org/pub/pdb/validation_reports/km/5kmw ftp://data.pdbj.org/pub/pdb/validation_reports/km/5kmw | HTTPS FTP |
-Related structure data
| Related structure data |  5u2x  5u2y  5u2z |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 27503.123 Da / Num. of mol.: 1 / Mutation: E165A, R271N, R273N Source method: isolated from a genetically manipulated source Source: (gene. exp.)   | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #2: Chemical | ChemComp-SO4 / #3: Chemical | ChemComp-PNM / | #4: Chemical | #5: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.69 Å3/Da / Density % sol: 54.29 % |
|---|---|
| Crystal grow | Temperature: 293.15 K / Method: batch mode / pH: 6.1 Details: 30 microliters of 10 mg/ml protein concentration was added to a solution containing 2.0 M ammonium sulfate and 0.1 M sodium citrate (pH 6.1). For ligand soaking, crystals were placed for 2-3 ...Details: 30 microliters of 10 mg/ml protein concentration was added to a solution containing 2.0 M ammonium sulfate and 0.1 M sodium citrate (pH 6.1). For ligand soaking, crystals were placed for 2-3 h in a reservoir solution containing 2.7 M ammonium sulfate, 0.1 M sodium citrate (pH 6.1), and 5.0 mM benzyl penicillin. The crystals were then placed momentarily in a reservoir solution containing a cryoprotectant (30% w/v trehalose) and subsequently flash-frozen in liquid nitrogen |
-Data collection
| Diffraction | Mean temperature: 15 K Ambient temp details: Cryo industries of America cryocool Helium cryostream |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 0.67 Å / Beamline: 19-ID / Wavelength: 0.67 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: May 1, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.67 Å / Relative weight: 1 |
| Reflection | Resolution: 1.1→38.62 Å / Num. obs: 118234 / % possible obs: 99.7 % / Redundancy: 5.5 % / Rmerge(I) obs: 0.093 / Net I/σ(I): 5.6 |
| Reflection shell | Resolution: 1.1→1.16 Å / Redundancy: 5.6 % / Rmerge(I) obs: 0.4 / Mean I/σ(I) obs: 2.1 / % possible all: 98.6 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.1→10 Å / Num. parameters: 23165 / Num. restraintsaints: 28142 / Cross valid method: FREE R / σ(F): 0 / Stereochemistry target values: ENGH AND HUBER MOLECULAR REPLACEMENT / Resolution: 1.1→10 Å / Num. parameters: 23165 / Num. restraintsaints: 28142 / Cross valid method: FREE R / σ(F): 0 / Stereochemistry target values: ENGH AND HUBERDetails: ANISOTROPIC REFINEMENT REDUCED FREE R (NO CUTOFF) BY ?
| |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 19 / Occupancy sum non hydrogen: 2443.8 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 1.1→10 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj