[English] 日本語
 Yorodumi
Yorodumi- PDB-5j7q: Macrophage Migration Inhibitory Factor bound to Inhibitor K664 De... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5j7q | ||||||
|---|---|---|---|---|---|---|---|
| Title | Macrophage Migration Inhibitory Factor bound to Inhibitor K664 Derivative | ||||||
 Components Components | Macrophage migration inhibitory factor | ||||||
 Keywords Keywords | Isomerase/Isomerase Inhibitor / Isomerase / Isomerase Inhibitor / Isomerase-Isomerase Inhibitor complex | ||||||
| Function / homology |  Function and homology information Function and homology information: / positive regulation of myeloid leukocyte cytokine production involved in immune response / phenylpyruvate tautomerase / L-dopachrome isomerase / dopachrome isomerase activity / phenylpyruvate tautomerase activity / positive regulation of arachidonate secretion / cytokine receptor binding / negative regulation of myeloid cell apoptotic process / negative regulation of macrophage chemotaxis ...: / positive regulation of myeloid leukocyte cytokine production involved in immune response / phenylpyruvate tautomerase / L-dopachrome isomerase / dopachrome isomerase activity / phenylpyruvate tautomerase activity / positive regulation of arachidonate secretion / cytokine receptor binding / negative regulation of myeloid cell apoptotic process / negative regulation of macrophage chemotaxis / negative regulation of mature B cell apoptotic process / carboxylic acid metabolic process / positive regulation of lipopolysaccharide-mediated signaling pathway / positive regulation of chemokine (C-X-C motif) ligand 2 production / prostaglandin biosynthetic process / negative regulation of protein metabolic process / negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / regulation of macrophage activation / chemoattractant activity / protein homotrimerization / negative regulation of DNA damage response, signal transduction by p53 class mediator / positive regulation of cAMP/PKA signal transduction / negative regulation of cellular senescence / positive regulation of phosphorylation / positive regulation of B cell proliferation / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / negative regulation of cell migration / cytokine activity / positive regulation of cytokine production / Cell surface interactions at the vascular wall / DNA damage response, signal transduction by p53 class mediator / positive regulation of fibroblast proliferation / positive regulation of tumor necrosis factor production / cellular senescence / protease binding / secretory granule lumen / vesicle / ficolin-1-rich granule lumen / cell surface receptor signaling pathway / positive regulation of ERK1 and ERK2 cascade / inflammatory response / negative regulation of gene expression / innate immune response / positive regulation of cell population proliferation / Neutrophil degranulation / negative regulation of apoptotic process / cell surface / extracellular space / extracellular exosome / extracellular region / nucleoplasm / identical protein binding / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.05 Å MOLECULAR REPLACEMENT / Resolution: 2.05 Å | ||||||
 Authors Authors | Robertson, M.J. / Jorgensen, W.L. | ||||||
 Citation Citation |  Journal: Bioorg.Med.Chem.Lett. / Year: 2016 Journal: Bioorg.Med.Chem.Lett. / Year: 2016Title: Irregularities in enzyme assays: The case of macrophage migration inhibitory factor. Authors: Cisneros, J.A. / Robertson, M.J. / Valhondo, M. / Jorgensen, W.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5j7q.cif.gz 5j7q.cif.gz | 83.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5j7q.ent.gz pdb5j7q.ent.gz | 61.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5j7q.json.gz 5j7q.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/j7/5j7q https://data.pdbj.org/pub/pdb/validation_reports/j7/5j7q ftp://data.pdbj.org/pub/pdb/validation_reports/j7/5j7q ftp://data.pdbj.org/pub/pdb/validation_reports/j7/5j7q | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5j7pC  3u18S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 3 molecules ABC
| #1: Protein | Mass: 12355.056 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: MIF, GLIF, MMIF / Production host: Homo sapiens (human) / Gene: MIF, GLIF, MMIF / Production host:  References: UniProt: P14174, phenylpyruvate tautomerase, L-dopachrome isomerase |
|---|
-Non-polymers , 5 types, 83 molecules 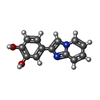








| #2: Chemical | ChemComp-6H2 / | ||||||
|---|---|---|---|---|---|---|---|
| #3: Chemical | ChemComp-SO4 / #4: Chemical | #5: Chemical | ChemComp-IPA / | #6: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.73 Å3/Da / Density % sol: 54.88 % |
|---|---|
| Crystal grow | Temperature: 293.15 K / Method: vapor diffusion, hanging drop / pH: 7 Details: 2.4 M Ammonium Sulfate, 3% isopropanol and 0.1 M Tris pH 7.0 |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RIGAKU SATURN 944+ / Detector: CCD / Date: Jul 15, 2015 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.95→53.71 Å / Num. obs: 29961 / % possible obs: 98.6 % / Redundancy: 6.3 % / Rmerge(I) obs: 0.088 / Rpim(I) all: 0.037 / Rrim(I) all: 0.095 / Χ2: 0.94 / Net I/av σ(I): 18.615 / Net I/σ(I): 6.4 / Num. measured all: 187842 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / Rejects: _
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3U18 Resolution: 2.05→53.71 Å / Cor.coef. Fo:Fc: 0.951 / Cor.coef. Fo:Fc free: 0.93 / SU B: 4.432 / SU ML: 0.118 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.206 / ESU R Free: 0.175 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : REFINED INDIVIDUALLY
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 104.01 Å2 / Biso mean: 27.31 Å2 / Biso min: 14.67 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.05→53.71 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.045→2.098 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







