[English] 日本語
 Yorodumi
Yorodumi- PDB-5j1o: Excited state (Bound-like) sampled during RDC restrained Replica-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5j1o | ||||||
|---|---|---|---|---|---|---|---|
| Title | Excited state (Bound-like) sampled during RDC restrained Replica-averaged Metadynamics (RAM) simulations of the HIV-1 TAR complexed with cyclic peptide mimetic of Tat | ||||||
 Components Components |
| ||||||
 Keywords Keywords | VIRAL PROTEIN / TAR:Tat complex / RAM simulations / Residual dipolar couplings / excited state | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||
| Method | SOLUTION NMR / molecular dynamics | ||||||
 Authors Authors | Borkar, A.N. / Bardaro, M.F. / Varani, G. / Vendruscolo, M. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2016 Journal: Proc.Natl.Acad.Sci.USA / Year: 2016Title: Structure of a low-population binding intermediate in protein-RNA recognition. Authors: Borkar, A.N. / Bardaro, M.F. / Camilloni, C. / Aprile, F.A. / Varani, G. / Vendruscolo, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5j1o.cif.gz 5j1o.cif.gz | 258.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5j1o.ent.gz pdb5j1o.ent.gz | 198.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5j1o.json.gz 5j1o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/j1/5j1o https://data.pdbj.org/pub/pdb/validation_reports/j1/5j1o ftp://data.pdbj.org/pub/pdb/validation_reports/j1/5j1o ftp://data.pdbj.org/pub/pdb/validation_reports/j1/5j1o | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 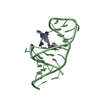
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 1768.189 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
|---|---|
| #2: RNA chain | Mass: 9307.555 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: HIV-1 TAR RNA / Source: (gene. exp.)   Human immunodeficiency virus 1 Human immunodeficiency virus 1Production host: in vitro transcription vector pT7-TP(deltai) (others) |
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR experiment | Sample state: anisotropic / Type: not applicable |
- Sample preparation
Sample preparation
| Details | Type: solution Contents: 2 uM [U-99% 13C; U-99% 15N] RNA and peptide, 100% D2O Label: peptide / Solvent system: 100% D2O |
|---|---|
| Sample | Conc.: 2 uM / Component: RNA and peptide / Isotopic labeling: [U-99% 13C; U-99% 15N] |
| Sample conditions | Ionic strength: 150 mM / Label: conditions1 / pH: 6.6 / Pressure: 1 atm / Temperature: 298.15 K |
-NMR measurement
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model: INOVA / Field strength: 800 MHz |
|---|
- Processing
Processing
| NMR software | Name:  Gromacs, Plumed / Developer: Vendruscolo, Camilloni and Borkar / Classification: structure calculation Gromacs, Plumed / Developer: Vendruscolo, Camilloni and Borkar / Classification: structure calculation |
|---|---|
| Refinement | Method: molecular dynamics / Software ordinal: 1 / Details: RDC restrained RAM simulations |
| NMR representative | Selection criteria: lowest energy |
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 7790 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller












 PDBj
PDBj





























