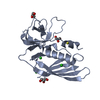
Entry Database : PDB / ID : 5d3iTitle Crystal structure of the SSL3-TLR2 complex Staphylococcal Superantigen-Like protein 3 Toll-like receptor 2 Keywords / / / / / / / / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Mus musculus (house mouse)Staphylococcus aureus (bacteria)Method / / / Resolution : 3.2 Å Authors Feitsma, L.J. / Huizinga, E.G. Funding support Organization Grant number Country Netherlands Organization for Scientific Research ECHO Grant 700.58.006 Netherlands Organization for Health Research and Development ZonMw Grant 205200004 Dutch Top Institute Pharma Project D1-101
Journal : Proc.Natl.Acad.Sci.USA / Year : 2015Title : Structural basis for inhibition of TLR2 by staphylococcal superantigen-like protein 3 (SSL3).Authors : Koymans, K.J. / Feitsma, L.J. / Brondijk, T.H. / Aerts, P.C. / Lukkien, E. / Lossl, P. / van Kessel, K.P. / de Haas, C.J. / van Strijp, J.A. / Huizinga, E.G. History Deposition Aug 6, 2015 Deposition site / Processing site Revision 1.0 Aug 19, 2015 Provider / Type Revision 1.1 Sep 2, 2015 Group Revision 1.2 Sep 9, 2015 Group Revision 2.0 Jul 29, 2020 Group Advisory / Atomic model ... Advisory / Atomic model / Data collection / Derived calculations / Structure summary Category atom_site / chem_comp ... atom_site / chem_comp / entity / pdbx_branch_scheme / pdbx_chem_comp_identifier / pdbx_entity_branch / pdbx_entity_branch_descriptor / pdbx_entity_branch_link / pdbx_entity_branch_list / pdbx_entity_nonpoly / pdbx_nonpoly_scheme / pdbx_struct_assembly_gen / pdbx_unobs_or_zero_occ_atoms / struct_asym / struct_conn / struct_site / struct_site_gen Item _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ... _atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _atom_site.auth_asym_id / _atom_site.auth_seq_id / _atom_site.label_asym_id / _atom_site.label_entity_id / _chem_comp.name / _chem_comp.type / _pdbx_entity_nonpoly.entity_id / _pdbx_entity_nonpoly.name / _pdbx_struct_assembly_gen.asym_id_list / _pdbx_unobs_or_zero_occ_atoms.label_asym_id / _struct_conn.pdbx_role / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id Description / Provider / Type Revision 2.1 May 1, 2024 Group Data collection / Database references ... Data collection / Database references / Refinement description / Structure summary Category chem_comp / chem_comp_atom ... chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_DOI / _database_2.pdbx_database_accessionRevision 2.2 Oct 9, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information

 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.2 Å
MOLECULAR REPLACEMENT / Resolution: 3.2 Å  Authors
Authors Netherlands, 3items
Netherlands, 3items  Citation
Citation Journal: Proc.Natl.Acad.Sci.USA / Year: 2015
Journal: Proc.Natl.Acad.Sci.USA / Year: 2015 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5d3i.cif.gz
5d3i.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5d3i.ent.gz
pdb5d3i.ent.gz PDB format
PDB format 5d3i.json.gz
5d3i.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 5d3i_validation.pdf.gz
5d3i_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 5d3i_full_validation.pdf.gz
5d3i_full_validation.pdf.gz 5d3i_validation.xml.gz
5d3i_validation.xml.gz 5d3i_validation.cif.gz
5d3i_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/d3/5d3i
https://data.pdbj.org/pub/pdb/validation_reports/d3/5d3i ftp://data.pdbj.org/pub/pdb/validation_reports/d3/5d3i
ftp://data.pdbj.org/pub/pdb/validation_reports/d3/5d3i Links
Links Assembly
Assembly
 Components
Components
 Homo sapiens (human) / References: UniProt: Q9QUN7
Homo sapiens (human) / References: UniProt: Q9QUN7 Staphylococcus aureus (strain NCTC 8325) (bacteria)
Staphylococcus aureus (strain NCTC 8325) (bacteria)




 X-RAY DIFFRACTION
X-RAY DIFFRACTION Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  PETRA III, DESY
PETRA III, DESY  / Beamline: P11 / Wavelength: 1 Å
/ Beamline: P11 / Wavelength: 1 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj















