+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4zkc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
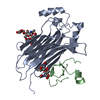
| Title | The chemokine binding protein of orf virus complexed with CCL7 | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | Viral Protein/cytokine / Complex / Orf virus chemokine binding protein / CCL7 / Viral Protein-cytokine complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationCCR2 chemokine receptor binding / CCR1 chemokine receptor binding / positive regulation of natural killer cell chemotaxis / CCR chemokine receptor binding / chemokine-mediated signaling pathway / eosinophil chemotaxis / chemokine activity / Chemokine receptors bind chemokines / monocyte chemotaxis / cellular response to ethanol ...CCR2 chemokine receptor binding / CCR1 chemokine receptor binding / positive regulation of natural killer cell chemotaxis / CCR chemokine receptor binding / chemokine-mediated signaling pathway / eosinophil chemotaxis / chemokine activity / Chemokine receptors bind chemokines / monocyte chemotaxis / cellular response to ethanol / cytoskeleton organization / response to gamma radiation / intracellular calcium ion homeostasis / chemotaxis / antimicrobial humoral immune response mediated by antimicrobial peptide / cell-cell signaling / regulation of cell shape / heparin binding / positive regulation of cell migration / inflammatory response / signal transduction / extracellular space / extracellular region / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Orf virus Orf virus Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.15 Å MOLECULAR REPLACEMENT / Resolution: 3.15 Å | |||||||||
 Authors Authors | Knapp, K.M. / Nakatani, Y. / Krause, K.L. | |||||||||
 Citation Citation |  Journal: Structure / Year: 2015 Journal: Structure / Year: 2015Title: Structures of Orf Virus Chemokine Binding Protein in Complex with Host Chemokines Reveal Clues to Broad Binding Specificity. Authors: Counago, R.M. / Knapp, K.M. / Nakatani, Y. / Fleming, S.B. / Corbett, M. / Wise, L.M. / Mercer, A.A. / Krause, K.L. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4zkc.cif.gz 4zkc.cif.gz | 116.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4zkc.ent.gz pdb4zkc.ent.gz | 87.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4zkc.json.gz 4zkc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4zkc_validation.pdf.gz 4zkc_validation.pdf.gz | 975 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4zkc_full_validation.pdf.gz 4zkc_full_validation.pdf.gz | 978.5 KB | Display | |
| Data in XML |  4zkc_validation.xml.gz 4zkc_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  4zkc_validation.cif.gz 4zkc_validation.cif.gz | 15.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zk/4zkc https://data.pdbj.org/pub/pdb/validation_reports/zk/4zkc ftp://data.pdbj.org/pub/pdb/validation_reports/zk/4zkc ftp://data.pdbj.org/pub/pdb/validation_reports/zk/4zkc | HTTPS FTP |
-Related structure data
| Related structure data |  4p5iSC  4zk9C  4zkbC C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 30374.613 Da / Num. of mol.: 1 / Fragment: UNP residues 17-286 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Orf virus (strain NZ2) / Plasmid: PTT5 / Cell line (production host): HEK 293-6E / Production host: Orf virus (strain NZ2) / Plasmid: PTT5 / Cell line (production host): HEK 293-6E / Production host:  Homo sapiens (human) / References: UniProt: Q2F862 Homo sapiens (human) / References: UniProt: Q2F862 |
|---|---|
| #2: Protein | Mass: 9803.363 Da / Num. of mol.: 1 / Fragment: UNP residues 24-99 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CCL7, MCP3, SCYA6, SCYA7 / Plasmid: pTT5 / Cell line (production host): HEK 293-6E / Production host: Homo sapiens (human) / Gene: CCL7, MCP3, SCYA6, SCYA7 / Plasmid: pTT5 / Cell line (production host): HEK 293-6E / Production host:  Homo sapiens (human) / References: UniProt: P80098 Homo sapiens (human) / References: UniProt: P80098 |
| #3: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
| #4: Polysaccharide | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta- ...beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.39 Å3/Da / Density % sol: 63.68 % |
|---|---|
| Crystal grow | Temperature: 289 K / Method: vapor diffusion, hanging drop / pH: 5.6 Details: 0.2M potassium sodium tartrate tetrahydrate, 0.1M sodium citrate tribasic dihydrate pH5.6, 2.0M ammonium citrate |
-Data collection
| Diffraction | Mean temperature: 93.2 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Australian Synchrotron Australian Synchrotron  / Beamline: MX1 / Wavelength: 0.95369 Å / Beamline: MX1 / Wavelength: 0.95369 Å |
| Detector | Type: ADSC QUANTUM 210r / Detector: CCD / Date: Aug 26, 2011 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.95369 Å / Relative weight: 1 |
| Reflection | Resolution: 3.15→71.22 Å / Num. obs: 9921 / % possible obs: 97.4 % / Redundancy: 7.4 % / Biso Wilson estimate: 85.1 Å2 / Rmerge(I) obs: 0.082 / Net I/σ(I): 15.7 |
| Reflection shell | Resolution: 3.15→3.32 Å / Redundancy: 4.6 % / Rmerge(I) obs: 0.596 / Mean I/σ(I) obs: 2.3 / % possible all: 82.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4P5I Resolution: 3.15→71.22 Å / Cor.coef. Fo:Fc: 0.896 / Cor.coef. Fo:Fc free: 0.845 / SU B: 50.881 / SU ML: 0.42 / Cross valid method: THROUGHOUT / ESU R: 0.849 / ESU R Free: 0.475 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 102.438 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.15→71.22 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj