| Entry | Database: PDB / ID: 4zhu
|
|---|
| Title | Crystal structure of a bacterial repressor protein |
|---|
 Components Components | YfiR |
|---|
 Keywords Keywords | TRANSCRIPTION / Periplasmic binding protein / repressor |
|---|
| Function / homology | YfiR/HmsC-like / YfiR/HmsC-like / periplasmic space / Negative regulator YfiR Function and homology information Function and homology information |
|---|
| Biological species |  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.3968 Å SYNCHROTRON / Resolution: 2.3968 Å |
|---|
 Authors Authors | Li, S. / Li, T. / Wang, Y. / Bartlam, M. |
|---|
| Funding support |  China, 4items China, 4items | Organization | Grant number | Country |
|---|
| Ministry of Science & Technology | 2014CB560709 |  China China | | National Science Foundation of China | 31450110071 |  China China | | National Science Foundation of China | 31322012 |  China China | | Tianjin Municipal Science and Technology Commission | 13JCYBJC20800 |  China China |
|
|---|
 Citation Citation |  Journal: Sci Rep / Year: 2015 Journal: Sci Rep / Year: 2015
Title: Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Authors: Li, S. / Li, T. / Xu, Y. / Zhang, Q. / Zhang, W. / Che, S. / Liu, R. / Wang, Y. / Bartlam, M. |
|---|
| History | | Deposition | Apr 27, 2015 | Deposition site: RCSB / Processing site: PDBJ |
|---|
| Revision 1.0 | Apr 27, 2016 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Nov 20, 2024 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Structure summary
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature / pdbx_struct_oper_list
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_oper_list.symmetry_operation |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Pseudomonas aeruginosa PAO1 (bacteria)
Pseudomonas aeruginosa PAO1 (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.3968 Å
SYNCHROTRON / Resolution: 2.3968 Å  Authors
Authors China, 4items
China, 4items  Citation
Citation Journal: Sci Rep / Year: 2015
Journal: Sci Rep / Year: 2015 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4zhu.cif.gz
4zhu.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4zhu.ent.gz
pdb4zhu.ent.gz PDB format
PDB format 4zhu.json.gz
4zhu.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/zh/4zhu
https://data.pdbj.org/pub/pdb/validation_reports/zh/4zhu ftp://data.pdbj.org/pub/pdb/validation_reports/zh/4zhu
ftp://data.pdbj.org/pub/pdb/validation_reports/zh/4zhu Links
Links Assembly
Assembly
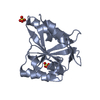

 Components
Components Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 / Gene: yfiR / Production host:
Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 / Gene: yfiR / Production host: 
 X-RAY DIFFRACTION
X-RAY DIFFRACTION Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRF
SSRF  / Beamline: BL17U / Wavelength: 1 Å
/ Beamline: BL17U / Wavelength: 1 Å Processing
Processing Movie
Movie Controller
Controller
















 PDBj
PDBj

