[English] 日本語
 Yorodumi
Yorodumi- PDB-4xdv: Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4xdv | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclohexanediol | ||||||
 Components Components | Limonene-1,2-epoxide hydrolase | ||||||
 Keywords Keywords | HYDROLASE / epoxide hydrolase | ||||||
| Function / homology |  Function and homology information Function and homology informationlimonene-1,2-epoxide hydrolase / limonene-1,2-epoxide hydrolase activity Similarity search - Function | ||||||
| Biological species |  Rhodococcus erythropolis (bacteria) Rhodococcus erythropolis (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.25 Å molecular replacement / Resolution: 2.25 Å | ||||||
| Model details | A mutant of limonene 1,2-epoxide hydrolase | ||||||
 Authors Authors | Kong, X.D. / Sun, Z. / Lonsdale, R. / Xu, J.H. / Reetz, M.T. / Zhou, J. | ||||||
 Citation Citation |  Journal: Angew.Chem.Int.Ed.Engl. / Year: 2015 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2015Title: Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution Authors: Sun, Z. / Lonsdale, R. / Kong, X.D. / Xu, J.H. / Zhou, J. / Reetz, M.T. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4xdv.cif.gz 4xdv.cif.gz | 243 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4xdv.ent.gz pdb4xdv.ent.gz | 197.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4xdv.json.gz 4xdv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4xdv_validation.pdf.gz 4xdv_validation.pdf.gz | 518.5 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4xdv_full_validation.pdf.gz 4xdv_full_validation.pdf.gz | 532.3 KB | Display | |
| Data in XML |  4xdv_validation.xml.gz 4xdv_validation.xml.gz | 48.9 KB | Display | |
| Data in CIF |  4xdv_validation.cif.gz 4xdv_validation.cif.gz | 69.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xd/4xdv https://data.pdbj.org/pub/pdb/validation_reports/xd/4xdv ftp://data.pdbj.org/pub/pdb/validation_reports/xd/4xdv ftp://data.pdbj.org/pub/pdb/validation_reports/xd/4xdv | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological unit is a dimer. There are 2 biological units in the asymmetric unit (chains A & B and chains C & D) |
- Components
Components
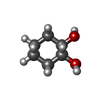
| #1: Protein | Mass: 17369.381 Da / Num. of mol.: 8 / Fragment: UNP residues 5-149 / Mutation: L74F/M78V/I80V/L114F Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Rhodococcus erythropolis (bacteria) / Gene: limA / Plasmid: pET22b / Production host: Rhodococcus erythropolis (bacteria) / Gene: limA / Plasmid: pET22b / Production host:  #2: Chemical | ChemComp-40O / ( #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.97 Å3/Da / Density % sol: 58.59 % Description: THE ENTRY CONTAINS FRIEDEL PAIRS IN F_PLUS/MINUS COLUMNS. |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, sitting drop / pH: 6.5 / Details: 1.4 M citrate sodium |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RIGAKU RAXIS IV++ / Detector: IMAGE PLATE / Date: Oct 25, 2014 / Details: mirrors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: GRAPHITE / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.25→50 Å / Num. obs: 70793 / % possible obs: 97.6 % / Redundancy: 5.3 % / Biso Wilson estimate: 41.26 Å2 / Rmerge(I) obs: 0.114 / Rpim(I) all: 0.055 / Rrim(I) all: 0.127 / Χ2: 1.001 / Net I/av σ(I): 10.503 / Net I/σ(I): 11.6 / Num. measured all: 376437 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / Rejects: _
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.25→33.524 Å / FOM work R set: 0.7422 / SU ML: 0.31 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 31.97 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 2.25→33.524 Å / FOM work R set: 0.7422 / SU ML: 0.31 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 31.97 / Stereochemistry target values: MLDetails: SF FILE CONTAINS FRIEDEL PAIRS UNDER I/F_MINUS AND I/F_PLUS COLUMNS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 90.18 Å2 / Biso mean: 33.96 Å2 / Biso min: 8.91 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.25→33.524 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 25
|
 Movie
Movie Controller
Controller















 PDBj
PDBj