[English] 日本語
 Yorodumi
Yorodumi- PDB-4x6o: FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl (4-{4-chloro-2-[(... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4x6o | ||||||
|---|---|---|---|---|---|---|---|
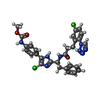
| Title | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl (4-{4-chloro-2-[(1S)-1-({3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]propanoyl}amino)-2-phenylethyl]-1H-imidazol-5-yl}phenyl)carbamate | ||||||
 Components Components | Coagulation factor XI, light chain | ||||||
 Keywords Keywords | hydrolase/hydrolase inhibitor / SERINE PROTEASE / BLOOD COAGULATION FACTOR / PROTEIN INHIBITOR COMPLEX / HYDROLASE-HYDROLASE INHIBITOR COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationcoagulation factor XIa / serine-type aminopeptidase activity / Defective F9 activation / positive regulation of fibrinolysis / plasminogen activation / Intrinsic Pathway of Fibrin Clot Formation / blood coagulation / heparin binding / serine-type endopeptidase activity / extracellular space ...coagulation factor XIa / serine-type aminopeptidase activity / Defective F9 activation / positive regulation of fibrinolysis / plasminogen activation / Intrinsic Pathway of Fibrin Clot Formation / blood coagulation / heparin binding / serine-type endopeptidase activity / extracellular space / extracellular exosome / extracellular region / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | ||||||
 Authors Authors | Wei, A. | ||||||
 Citation Citation |  Journal: Bioorg.Med.Chem.Lett. / Year: 2015 Journal: Bioorg.Med.Chem.Lett. / Year: 2015Title: Structure-based design of inhibitors of coagulation factor XIa with novel P1 moieties. Authors: Pinto, D.J. / Smallheer, J.M. / Corte, J.R. / Austin, E.J. / Wang, C. / Fang, T. / Smith, L.M. / Rossi, K.A. / Rendina, A.R. / Bozarth, J.M. / Zhang, G. / Wei, A. / Ramamurthy, V. / Sheriff, ...Authors: Pinto, D.J. / Smallheer, J.M. / Corte, J.R. / Austin, E.J. / Wang, C. / Fang, T. / Smith, L.M. / Rossi, K.A. / Rendina, A.R. / Bozarth, J.M. / Zhang, G. / Wei, A. / Ramamurthy, V. / Sheriff, S. / Myers, J.E. / Morin, P.E. / Luettgen, J.M. / Seiffert, D.A. / Quan, M.L. / Wexler, R.R. #1:  Journal: J.Med.Chem. / Year: 2014 Journal: J.Med.Chem. / Year: 2014Title: Tetrahydroquinoline derivatives as potent and selective factor XIa inhibitors. Authors: Quan, M.L. / Wong, P.C. / Wang, C. / Woerner, F. / Smallheer, J.M. / Barbera, F.A. / Bozarth, J.M. / Brown, R.L. / Harpel, M.R. / Luettgen, J.M. / Morin, P.E. / Peterson, T. / Ramamurthy, V. ...Authors: Quan, M.L. / Wong, P.C. / Wang, C. / Woerner, F. / Smallheer, J.M. / Barbera, F.A. / Bozarth, J.M. / Brown, R.L. / Harpel, M.R. / Luettgen, J.M. / Morin, P.E. / Peterson, T. / Ramamurthy, V. / Rendina, A.R. / Rossi, K.A. / Watson, C.A. / Wei, A. / Zhang, G. / Seiffert, D. / Wexler, R.R. #2:  Journal: J.Med.Chem. / Year: 2014 Journal: J.Med.Chem. / Year: 2014Title: Phenylimidazoles as Potent and Selective Inhibitors of Coagulation Factor XIa with in Vivo Antithrombotic Activity. Authors: Hangeland, J.J. / Friends, T.J. / Rossi, K.A. / Smallheer, J.M. / Wang, C. / Sun, Z. / Corte, J.R. / Fang, T. / Wong, P.C. / Rendina, A.R. / Barbera, F.A. / Bozarth, J.M. / Luettgen, J.M. / ...Authors: Hangeland, J.J. / Friends, T.J. / Rossi, K.A. / Smallheer, J.M. / Wang, C. / Sun, Z. / Corte, J.R. / Fang, T. / Wong, P.C. / Rendina, A.R. / Barbera, F.A. / Bozarth, J.M. / Luettgen, J.M. / Watson, C.A. / Zhang, G. / Wei, A. / Ramamurthy, V. / Morin, P.E. / Bisacchi, G.S. / Subramaniam, S. / Arunachalam, P. / Mathur, A. / Seiffert, D.A. / Wexler, R.R. / Quan, M.L. #3:  Journal: Bioorg.Med.Chem.Lett. / Year: 2015 Journal: Bioorg.Med.Chem.Lett. / Year: 2015Title: Pyridine and pyridinone-based factor XIa inhibitors. Authors: Corte, J.R. / Fang, T. / Hangeland, J.J. / Friends, T.J. / Rendina, A.R. / Luettgen, J.M. / Bozarth, J.M. / Barbera, F.A. / Rossi, K.A. / Wei, A. / Ramamurthy, V. / Morin, P.E. / Seiffert, D. ...Authors: Corte, J.R. / Fang, T. / Hangeland, J.J. / Friends, T.J. / Rendina, A.R. / Luettgen, J.M. / Bozarth, J.M. / Barbera, F.A. / Rossi, K.A. / Wei, A. / Ramamurthy, V. / Morin, P.E. / Seiffert, D.A. / Wexler, R.R. / Quan, M.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4x6o.cif.gz 4x6o.cif.gz | 71.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4x6o.ent.gz pdb4x6o.ent.gz | 50.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4x6o.json.gz 4x6o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4x6o_validation.pdf.gz 4x6o_validation.pdf.gz | 741.6 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4x6o_full_validation.pdf.gz 4x6o_full_validation.pdf.gz | 742.9 KB | Display | |
| Data in XML |  4x6o_validation.xml.gz 4x6o_validation.xml.gz | 13.7 KB | Display | |
| Data in CIF |  4x6o_validation.cif.gz 4x6o_validation.cif.gz | 19.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/x6/4x6o https://data.pdbj.org/pub/pdb/validation_reports/x6/4x6o ftp://data.pdbj.org/pub/pdb/validation_reports/x6/4x6o ftp://data.pdbj.org/pub/pdb/validation_reports/x6/4x6o | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
| |||||||||
| Details | chain A |
- Components
Components
| #1: Antibody | Mass: 27600.342 Da / Num. of mol.: 1 / Mutation: N113G, T115G Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: F11 / Plasmid: PET14B / Production host: Homo sapiens (human) / Gene: F11 / Plasmid: PET14B / Production host:  | ||||||
|---|---|---|---|---|---|---|---|
| #2: Chemical | ChemComp-3Y4 / | ||||||
| #3: Chemical | | #4: Chemical | ChemComp-EDO / #5: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.55 Å3/Da / Density % sol: 65.35 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 4.6 Details: 100 MM SODIUM ACETATE, PH 4.6, 26%(w/v) MEPEG2000, 200 MM AMMONIUM SULFATE |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU FR-E SUPERBRIGHT / Wavelength: 1.54 Å ROTATING ANODE / Type: RIGAKU FR-E SUPERBRIGHT / Wavelength: 1.54 Å |
| Detector | Type: RIGAKU SATURN 92 / Detector: CCD / Date: Aug 24, 2006 / Details: MICROMAX CONFOCAL |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54 Å / Relative weight: 1 |
| Reflection | Resolution: 2.1→50 Å / Num. obs: 22771 / % possible obs: 99.9 % / Observed criterion σ(I): 0 / Redundancy: 10.2 % / Biso Wilson estimate: 27.25 Å2 / Rmerge(I) obs: 0.146 / Net I/σ(I): 18 |
| Reflection shell | Resolution: 2.1→2.18 Å / Redundancy: 7.6 % / Rmerge(I) obs: 0.477 / Mean I/σ(I) obs: 3.7 / Rejects: 0 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: FACTOR XIA DOUBLE MUTANT Resolution: 2.1→26.33 Å / Cor.coef. Fo:Fc: 0.9424 / Cor.coef. Fo:Fc free: 0.9186 / SU R Cruickshank DPI: 0.152 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.163 / SU Rfree Blow DPI: 0.152 / SU Rfree Cruickshank DPI: 0.147
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 104.84 Å2 / Biso mean: 27.25 Å2 / Biso min: 11.33 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.228 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.1→26.33 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.1→2.2 Å / Total num. of bins used: 11
|
 Movie
Movie Controller
Controller















 PDBj
PDBj