[English] 日本語
 Yorodumi
Yorodumi- PDB-4uiw: BROMODOMAIN OF HUMAN BRD9 WITH N-(1,1-dioxo-1-thian-4-yl)-5-ethyl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4uiw | ||||||
|---|---|---|---|---|---|---|---|
| Title | BROMODOMAIN OF HUMAN BRD9 WITH N-(1,1-dioxo-1-thian-4-yl)-5-ethyl-4- oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2- carboximidamide | ||||||
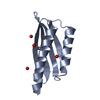
 Components Components | BROMODOMAIN-CONTAINING PROTEIN 9 | ||||||
 Keywords Keywords | TRANSCRIPTION / INHIBITOR / HISTONE / EPIGENETIC READER | ||||||
| Function / homology |  Function and homology information Function and homology informationFormation of the non-canonical BAF (ncBAF) complex / GBAF complex / : / SWI/SNF complex / positive regulation of stem cell population maintenance / histone reader activity / negative regulation of cell differentiation / double-strand break repair via homologous recombination / site of double-strand break / protein-macromolecule adaptor activity ...Formation of the non-canonical BAF (ncBAF) complex / GBAF complex / : / SWI/SNF complex / positive regulation of stem cell population maintenance / histone reader activity / negative regulation of cell differentiation / double-strand break repair via homologous recombination / site of double-strand break / protein-macromolecule adaptor activity / nucleic acid binding / chromatin remodeling / positive regulation of cell population proliferation / regulation of transcription by RNA polymerase II / chromatin / nucleoplasm / nucleus Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.73 Å MOLECULAR REPLACEMENT / Resolution: 1.73 Å | ||||||
 Authors Authors | Chung, C. / Theodoulou, N.T. / Bamborough, P. / Humphreys, P.G. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2016 Journal: J.Med.Chem. / Year: 2016Title: The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition. Authors: Theodoulou, N.H. / Bamborough, P. / Bannister, A.J. / Becher, I. / Bit, R.A. / Che, K.H. / Chung, C. / Dittmann, A. / Drewes, G. / Drewry, D.H. / Gordon, L. / Grandi, P. / Leveridge, M. / ...Authors: Theodoulou, N.H. / Bamborough, P. / Bannister, A.J. / Becher, I. / Bit, R.A. / Che, K.H. / Chung, C. / Dittmann, A. / Drewes, G. / Drewry, D.H. / Gordon, L. / Grandi, P. / Leveridge, M. / Lindon, M. / Michon, A. / Molnar, J. / Robson, S.C. / Tomkinson, N.C.O. / Kouzarides, T. / Prinjha, R.K. / Humphreys, P.G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4uiw.cif.gz 4uiw.cif.gz | 60.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4uiw.ent.gz pdb4uiw.ent.gz | 44.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4uiw.json.gz 4uiw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ui/4uiw https://data.pdbj.org/pub/pdb/validation_reports/ui/4uiw ftp://data.pdbj.org/pub/pdb/validation_reports/ui/4uiw ftp://data.pdbj.org/pub/pdb/validation_reports/ui/4uiw | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4uitC  4uiuC  4uivC  4uixC  4uiyC  4uizC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 12228.203 Da / Num. of mol.: 1 / Fragment: RESIDUES 133-238 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Production host: HOMO SAPIENS (human) / Production host:  |
|---|---|
| #2: Chemical | ChemComp-EDO / |
| #3: Chemical | ChemComp-H1B / |
| #4: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.38 Å3/Da / Density % sol: 48.29 % / Description: NONE |
|---|---|
| Crystal grow | Temperature: 277 K / pH: 6.5 Details: 0.1M MORPHEUS BUFFER PH 6.5, 30% MORPHEUS_EDO_P8K, 0.1M MORPHEUS AMINO ACIDS AT 4C |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Wavelength: 1.54178 ROTATING ANODE / Wavelength: 1.54178 |
| Detector | Date: Jan 29, 2014 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54178 Å / Relative weight: 1 |
| Reflection | Resolution: 1.73→31 Å / Num. obs: 10779 / % possible obs: 92.1 % / Observed criterion σ(I): 2 / Redundancy: 2.2 % / Rmerge(I) obs: 0.02 / Net I/σ(I): 26.1 |
| Reflection shell | Resolution: 1.73→1.83 Å / Redundancy: 1.9 % / Rmerge(I) obs: 0.07 / Mean I/σ(I) obs: 7.6 / % possible all: 85.3 |
- Processing
Processing
| Software | Name: REFMAC / Version: 5.6.0117 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: NONE Resolution: 1.73→31 Å / Cor.coef. Fo:Fc: 0.971 / Cor.coef. Fo:Fc free: 0.949 / SU B: 3.561 / SU ML: 0.064 / Cross valid method: THROUGHOUT / ESU R: 0.108 / ESU R Free: 0.108 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 17.298 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.73→31 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj