+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4qa0 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of C153F HDAC8 in complex with SAHA | ||||||
 Components Components | Histone deacetylase 8 | ||||||
 Keywords Keywords | HYDROLASE / metalloenzyme / histone deacetylase / enzyme inhibitor complex / Cornelia de Lange Syndrome / arginase/deacetylase fold | ||||||
| Function / homology |  Function and homology information Function and homology informationhistone decrotonylase activity / histone deacetylase activity, hydrolytic mechanism / histone deacetylase / protein lysine deacetylase activity / Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In linear amides / histone deacetylase activity / regulation of telomere maintenance / mitotic sister chromatid cohesion / Notch-HLH transcription pathway / nuclear chromosome ...histone decrotonylase activity / histone deacetylase activity, hydrolytic mechanism / histone deacetylase / protein lysine deacetylase activity / Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In linear amides / histone deacetylase activity / regulation of telomere maintenance / mitotic sister chromatid cohesion / Notch-HLH transcription pathway / nuclear chromosome / histone deacetylase complex / negative regulation of protein ubiquitination / Hsp70 protein binding / Resolution of Sister Chromatid Cohesion / HDACs deacetylate histones / Hsp90 protein binding / regulation of protein stability / NOTCH1 Intracellular Domain Regulates Transcription / Constitutive Signaling by NOTCH1 PEST Domain Mutants / Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants / Separation of Sister Chromatids / heterochromatin formation / chromatin organization / DNA-binding transcription factor binding / negative regulation of transcription by RNA polymerase II / nucleoplasm / metal ion binding / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.242 Å MOLECULAR REPLACEMENT / Resolution: 2.242 Å | ||||||
 Authors Authors | Decroos, C. / Bowman, C.B. / Moser, J.-A.S. / Christianson, K.E. / Deardorff, M.A. / Christianson, D.W. | ||||||
 Citation Citation |  Journal: Acs Chem.Biol. / Year: 2014 Journal: Acs Chem.Biol. / Year: 2014Title: Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders. Authors: Decroos, C. / Bowman, C.M. / Moser, J.A. / Christianson, K.E. / Deardorff, M.A. / Christianson, D.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4qa0.cif.gz 4qa0.cif.gz | 166.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4qa0.ent.gz pdb4qa0.ent.gz | 129.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4qa0.json.gz 4qa0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qa/4qa0 https://data.pdbj.org/pub/pdb/validation_reports/qa/4qa0 ftp://data.pdbj.org/pub/pdb/validation_reports/qa/4qa0 ftp://data.pdbj.org/pub/pdb/validation_reports/qa/4qa0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4qa1C  4qa2C  4qa3C  4qa4C  4qa5C  4qa6C  4qa7C  3ewfS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 43276.023 Da / Num. of mol.: 2 / Mutation: C153F Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: HDAC8, HDACL1, CDA07 / Plasmid: pHD2-Xa-His / Production host: Homo sapiens (human) / Gene: HDAC8, HDACL1, CDA07 / Plasmid: pHD2-Xa-His / Production host:  |
|---|
-Non-polymers , 5 types, 448 molecules 

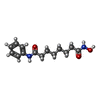






| #2: Chemical | | #3: Chemical | ChemComp-K / #4: Chemical | #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.46 Å3/Da / Density % sol: 49.94 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, sitting drop / pH: 8 Details: 0.1 M tris(hydroxymethyl)aminomethane) (TRIS, pH = 8), 4 mM tris(2-carboxyethyl)phosphine (TCEP), 17% PEG 8000, VAPOR DIFFUSION, SITTING DROP, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X29A / Wavelength: 1.075 Å / Beamline: X29A / Wavelength: 1.075 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Feb 23, 2014 / Details: mirrors |
| Radiation | Monochromator: SAGITALLY FOCUSED Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.075 Å / Relative weight: 1 |
| Reflection | Resolution: 2.24→50 Å / Num. all: 40284 / Num. obs: 40281 / % possible obs: 100 % / Observed criterion σ(F): 0 / Observed criterion σ(I): -3 / Redundancy: 7.5 % / Biso Wilson estimate: 28.65 Å2 / Rmerge(I) obs: 0.13 / Net I/σ(I): 13.7 |
| Reflection shell | Resolution: 2.24→2.32 Å / Redundancy: 7.2 % / Rmerge(I) obs: 0.711 / Mean I/σ(I) obs: 3 / Num. unique all: 4003 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 3EWF Resolution: 2.242→44.827 Å / SU ML: 0.26 / Isotropic thermal model: isotropic / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 23.02 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 33.49 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.242→44.827 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj






