[English] 日本語
 Yorodumi
Yorodumi- PDB-4pm3: Structure of the double-stranded DNA binding type IV secretion pr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4pm3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
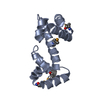
| Title | Structure of the double-stranded DNA binding type IV secretion protein TraN from Enterococcus | |||||||||
 Components Components | AM32 | |||||||||
 Keywords Keywords | DNA BINDING PROTEIN / Type IV secretion / internal dimer / isomerase | |||||||||
| Function / homology | isomerase activity / BROMIDE ION / : / AM32 Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.8 Å molecular replacement / Resolution: 1.8 Å | |||||||||
| Model details | helix-turn-helix-like motif, beta-barrel-like motif | |||||||||
 Authors Authors | Goessweiner-Mohr, N. / Keller, W. | |||||||||
 Citation Citation |  Journal: ACTA CRYSTALLOGR.,SECT.D / Year: 2014 Journal: ACTA CRYSTALLOGR.,SECT.D / Year: 2014Title: Structure of the double-stranded DNA binding type IV secretion protein TraN from Enterococcus Authors: Goessweiner-Mohr, N. / Eder, M. / Hofer, G. / Fercher, C. / Arends, K. / Birner-Gruenberger, R. / Grohmann, E. / Keller, W. #1: Journal: ACTA CRYSTALLOGR.,SECT.F / Year: 2012 Title: Crystallization and first data collection of the putative transfer protein TraN from Gram-positive conjugative plasmid pIP501 Authors: Goessweiner-Mohr, N. / Fercher, C. / Abajy, M.Y. / Grohmann, E. / Keller, W. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4pm3.cif.gz 4pm3.cif.gz | 68.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4pm3.ent.gz pdb4pm3.ent.gz | 48.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4pm3.json.gz 4pm3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pm/4pm3 https://data.pdbj.org/pub/pdb/validation_reports/pm/4pm3 ftp://data.pdbj.org/pub/pdb/validation_reports/pm/4pm3 ftp://data.pdbj.org/pub/pdb/validation_reports/pm/4pm3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4p0yC  4p0zSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological unit is a monomer. There are two biological units present in the asymmetric unit (chain A, chain B). |
- Components
Components
| #1: Protein | Mass: 17599.949 Da / Num. of mol.: 2 / Fragment: TraN Source method: isolated from a genetically manipulated source Details: MKHHHHHHHSDYDIPTTENLYFQGSGS is the 7x N-terminal HisTag HisTag and several N-terminal and C-terminal residues are not visible in the density map. Source: (gene. exp.)   #2: Chemical | #3: Chemical | ChemComp-BR / #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.93 Å3/Da / Density % sol: 36.16 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: microbatch / pH: 5.5 Details: 1:1 setup of purification buffer with Index screen condition 42 (0.1 M Bis-Tris, 25.0 % PEG 3350); final protein concentration: 3.25 mg/ml Crystal soaked for 1.5 h with Br4Pt |
-Data collection
| Diffraction | Mean temperature: 100 K / Ambient temp details: Pt LIII high-end remote | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 1.0615 Å / Beamline: X06DA / Wavelength: 1.0615 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: MARMOSAIC 225 mm CCD / Detector: CCD / Date: Apr 19, 2010 Details: toroidal mirror (M2) to vertically and horizontally focus the beam at the sample position (with 2:1 horizontal demagnification) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: BARTELS MONOCROMATOR / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.0615 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Redundancy: 3.6 % / Number: 69059 / Rsym value: 0.12 / D res high: 1.8 Å / D res low: 57.679 Å / Num. obs: 19191 / % possible obs: 100 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Diffraction reflection shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.8→57.68 Å / Num. all: 19191 / Num. obs: 19191 / % possible obs: 100 % / Redundancy: 3.6 % / Biso Wilson estimate: 14.79 Å2 / Rpim(I) all: 0.073 / Rrim(I) all: 0.141 / Rsym value: 0.12 / Net I/av σ(I): 4.67 / Net I/σ(I): 8.8 / Num. measured all: 69059 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / Rejects: _
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement | ||||||
|---|---|---|---|---|---|---|---|
| Phasing MR |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4P0Z Resolution: 1.8→28.967 Å / SU ML: 0.18 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 23.19 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.8→28.967 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj







