[English] 日本語
 Yorodumi
Yorodumi- PDB-4pd4: Structural analysis of atovaquone-inhibited cytochrome bc1 comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4pd4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural analysis of atovaquone-inhibited cytochrome bc1 complex reveals the molecular basis of antimalarial drug action | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | OXIDOREDUCTASE/INHIBITOR / Cytochrome bc1 complex / Membrane protein complex / antimalarial drug / inhibitor / OXIDOREDUCTASE-INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationComplex III assembly / : / matrix side of mitochondrial inner membrane / : / Respiratory electron transport / Mitochondrial protein degradation / mitochondrial respiratory chain complex III assembly / cellular respiration / respiratory chain complex III / quinol-cytochrome-c reductase ...Complex III assembly / : / matrix side of mitochondrial inner membrane / : / Respiratory electron transport / Mitochondrial protein degradation / mitochondrial respiratory chain complex III assembly / cellular respiration / respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / mitochondrial electron transport, ubiquinol to cytochrome c / mitochondrial crista / immunoglobulin complex / proton transmembrane transport / nuclear periphery / aerobic respiration / metalloendopeptidase activity / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / adaptive immune response / oxidoreductase activity / mitochondrial inner membrane / immune response / heme binding / mitochondrion / proteolysis / extracellular region / metal ion binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 3.04 Å molecular replacement / Resolution: 3.04 Å | |||||||||
 Authors Authors | Birth, D. / Kao, W.-C. / Hunte, C. | |||||||||
| Funding support |  Germany, 2items Germany, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2014 Journal: Nat Commun / Year: 2014Title: Structural analysis of atovaquone-inhibited cytochrome bc1 complex reveals the molecular basis of antimalarial drug action. Authors: Birth, D. / Kao, W.C. / Hunte, C. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4pd4.cif.gz 4pd4.cif.gz | 466.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4pd4.ent.gz pdb4pd4.ent.gz | 363.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4pd4.json.gz 4pd4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pd/4pd4 https://data.pdbj.org/pub/pdb/validation_reports/pd/4pd4 ftp://data.pdbj.org/pub/pdb/validation_reports/pd/4pd4 ftp://data.pdbj.org/pub/pdb/validation_reports/pd/4pd4 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Cytochrome b-c1 complex subunit ... , 7 types, 7 molecules ABEFGHI
| #1: Protein | Mass: 47445.242 Da / Num. of mol.: 1 / Fragment: UNP residues 27-457 / Source method: isolated from a natural source Source: (natural)  Strain: ATCC 204508 / S288c / References: UniProt: P07256 |
|---|---|
| #2: Protein | Mass: 38751.918 Da / Num. of mol.: 1 / Fragment: UNP residues 17-368 / Source method: isolated from a natural source Source: (natural)  Strain: ATCC 204508 / S288c / References: UniProt: P07257 |
| #5: Protein | Mass: 20122.955 Da / Num. of mol.: 1 / Fragment: UNP residues 31-215 / Source method: isolated from a natural source Source: (natural)  Strain: ATCC 204508 / S288c / References: UniProt: P08067, quinol-cytochrome-c reductase |
| #6: Protein | Mass: 8854.792 Da / Num. of mol.: 1 / Fragment: UNP residues 74-147 / Source method: isolated from a natural source Source: (natural)  Strain: ATCC 204508 / S288c / References: UniProt: P00127 |
| #7: Protein | Mass: 14452.557 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Strain: ATCC 204508 / S288c / References: UniProt: P00128 |
| #8: Protein | Mass: 10856.314 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Strain: ATCC 204508 / S288c / References: UniProt: P08525 |
| #9: Protein | Mass: 6535.484 Da / Num. of mol.: 1 / Fragment: UNP residues 2-58 / Source method: isolated from a natural source Source: (natural)  Strain: ATCC 204508 / S288c / References: UniProt: P22289 |
-Protein , 2 types, 2 molecules CD
| #3: Protein | Mass: 43686.590 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Strain: ATCC 204508 / S288c / References: UniProt: P00163 |
|---|---|
| #4: Protein | Mass: 27807.395 Da / Num. of mol.: 1 / Fragment: UNP residues 62-309 / Source method: isolated from a natural source Source: (natural)  Strain: ATCC 204508 / S288c / References: UniProt: P07143 |
-Antibody , 2 types, 2 molecules JK
| #10: Antibody | Mass: 14365.817 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #11: Antibody | Mass: 11926.350 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Non-polymers , 7 types, 11 molecules 


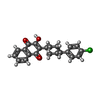









| #12: Chemical | ChemComp-UMQ / | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #13: Chemical | | #14: Chemical | #15: Chemical | ChemComp-AOQ / | #16: Chemical | ChemComp-3PE / | #17: Chemical | ChemComp-UQ6 / | #18: Chemical | ChemComp-FES / | |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.24 Å3/Da / Density % sol: 70.96 % |
|---|---|
| Crystal grow | Temperature: 277.2 K / Method: vapor diffusion, hanging drop / pH: 6.8 Details: polyethylene glycol 4000, DMSO, Sucrose, Potassium phosphate, n-undecyl-?-D-maltopyranoside, atovaquone |
-Data collection
| Diffraction | Mean temperature: 105 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 1 Å / Beamline: X06DA / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 2M-F / Detector: PIXEL / Date: Jun 25, 2012 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 3.04→25 Å / Num. obs: 77221 / % possible obs: 98.2 % / Redundancy: 6.8 % / Biso Wilson estimate: 97.24 Å2 / Net I/σ(I): 15.1 |
-Phasing
| Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR |
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 3.04→24.989 Å / SU ML: 0.52 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 39.2 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 3.04→24.989 Å / SU ML: 0.52 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 39.2 / Stereochemistry target values: MLDetails: Author states that the structure was determined by molecular replacement and used the high-resolution structure of yeast cyt bc1 complex (pdb 3cx5) as basis for refinement. Quality of x-ray ...Details: Author states that the structure was determined by molecular replacement and used the high-resolution structure of yeast cyt bc1 complex (pdb 3cx5) as basis for refinement. Quality of x-ray diffraction data for the new structure is limited and some poorly resolved loops were set to zero occupancy. The large complex covers a wide range of B-factors, with the highest in the matrix core subunits (chains A and B) and the lowest in the membrane integral subunit cytochrome b (chain C). In former structures obtained at room-temperature , such as 2KB9 at 2.3 angstrom, B factors range from above140 for chain A to below 30 for chain C. In this structure, the overall B-factors are lower but cover a similar range with very low up to zero B factors in the best ordered regions.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 130.91 Å2 / Biso mean: 45.1046 Å2 / Biso min: 0 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3.04→24.989 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 14
|
 Movie
Movie Controller
Controller










 PDBj
PDBj



















