[English] 日本語
 Yorodumi
Yorodumi- EMDB-8777: Pol I Initial Transcribing Complex with a truncated scaffold at 6... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8777 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pol I Initial Transcribing Complex with a truncated scaffold at 6.9 angstrom | ||||||||||||||||||
 Map data Map data | RNA polymerase I Initial Transcribing Complex with a truncated scaffold | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase I transcription regulatory region sequence-specific DNA binding / RNA polymerase I core factor complex / RNA polymerase I general transcription initiation factor activity / RNA polymerase I general transcription initiation factor binding / RNA polymerase I core promoter sequence-specific DNA binding / RNA polymerase I preinitiation complex assembly / RNA Polymerase I Transcription Initiation / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Pol II CTD phosphorylation and interaction with CE ...RNA polymerase I transcription regulatory region sequence-specific DNA binding / RNA polymerase I core factor complex / RNA polymerase I general transcription initiation factor activity / RNA polymerase I general transcription initiation factor binding / RNA polymerase I core promoter sequence-specific DNA binding / RNA polymerase I preinitiation complex assembly / RNA Polymerase I Transcription Initiation / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / termination of RNA polymerase III transcription / RNA Polymerase II Pre-transcription Events / RNA-templated transcription / Formation of TC-NER Pre-Incision Complex / regulation of cell size / RNA Polymerase I Promoter Escape / transcription initiation at RNA polymerase III promoter / termination of RNA polymerase I transcription / Gap-filling DNA repair synthesis and ligation in TC-NER / nucleolar large rRNA transcription by RNA polymerase I / transcription initiation at RNA polymerase I promoter / Estrogen-dependent gene expression / transcription by RNA polymerase III / Dual incision in TC-NER / RNA polymerase I complex / RNA polymerase III complex / transcription elongation by RNA polymerase I / RNA polymerase II, core complex / tRNA transcription by RNA polymerase III / transcription by RNA polymerase I / TBP-class protein binding / transcription initiation at RNA polymerase II promoter / transcription elongation by RNA polymerase II / promoter-specific chromatin binding / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / peroxisome / ribosome biogenesis / nucleic acid binding / transcription by RNA polymerase II / RNA polymerase II-specific DNA-binding transcription factor binding / protein dimerization activity / nucleolus / negative regulation of transcription by RNA polymerase II / mitochondrion / DNA binding / zinc ion binding / nucleoplasm / metal ion binding / nucleus / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
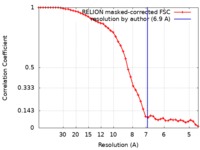
| Method | single particle reconstruction / cryo EM / Resolution: 6.9 Å | ||||||||||||||||||
 Authors Authors | Han Y / He Y | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Structural mechanism of ATP-independent transcription initiation by RNA polymerase I. Authors: Yan Han / Chunli Yan / Thi Hoang Duong Nguyen / Ashleigh J Jackobel / Ivaylo Ivanov / Bruce A Knutson / Yuan He /  Abstract: Transcription initiation by RNA Polymerase I (Pol I) depends on the Core Factor (CF) complex to recognize the upstream promoter and assemble into a Pre-Initiation Complex (PIC). Here, we solve a ...Transcription initiation by RNA Polymerase I (Pol I) depends on the Core Factor (CF) complex to recognize the upstream promoter and assemble into a Pre-Initiation Complex (PIC). Here, we solve a structure of Pol I-CF-DNA to 3.8 Å resolution using single-particle cryo-electron microscopy. The structure reveals a bipartite architecture of Core Factor and its recognition of the promoter from -27 to -16. Core Factor's intrinsic mobility correlates well with different conformational states of the Pol I cleft, in addition to the stabilization of either Rrn7 N-terminal domain near Pol I wall or the tandem winged helix domain of A49 at a partially overlapping location. Comparison of the three states in this study with the Pol II system suggests that a ratchet motion of the Core Factor-DNA sub-complex at upstream facilitates promoter melting in an ATP-independent manner, distinct from a DNA translocase actively threading the downstream DNA in the Pol II PIC. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8777.map.gz emd_8777.map.gz | 1.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8777-v30.xml emd-8777-v30.xml emd-8777.xml emd-8777.xml | 33.7 KB 33.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8777_fsc.xml emd_8777_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_8777.png emd_8777.png | 135.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8777 http://ftp.pdbj.org/pub/emdb/structures/EMD-8777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8777 | HTTPS FTP |
-Related structure data
| Related structure data |  8771C  8772C  8773C  8774C  8775C  8776C  5w5yC  5w64C  5w65C  5w66C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8777.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8777.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RNA polymerase I Initial Transcribing Complex with a truncated scaffold | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : RNA Polymerase I Initial Transcribing Complex with a truncated sc...
+Supramolecule #1: RNA Polymerase I Initial Transcribing Complex with a truncated sc...
+Macromolecule #1: DNA-directed RNA polymerase I subunit RPA190
+Macromolecule #2: DNA-directed RNA polymerase I subunit RPA135
+Macromolecule #3: DNA-directed RNA polymerases I and III subunit RPAC1
+Macromolecule #4: DNA-directed RNA polymerase I subunit RPA14
+Macromolecule #5: DNA-directed RNA polymerases I, II, and III subunit RPABC1
+Macromolecule #6: DNA-directed RNA polymerases I, II, and III subunit RPABC2
+Macromolecule #7: DNA-directed RNA polymerase I subunit RPA43
+Macromolecule #8: DNA-directed RNA polymerases I, II, and III subunit RPABC3
+Macromolecule #9: DNA-directed RNA polymerase I subunit RPA12
+Macromolecule #10: DNA-directed RNA polymerases I, II, and III subunit RPABC5
+Macromolecule #11: DNA-directed RNA polymerases I and III subunit RPAC2
+Macromolecule #12: DNA-directed RNA polymerases I, II, and III subunit RPABC4
+Macromolecule #13: DNA-directed RNA polymerase I subunit RPA49
+Macromolecule #14: DNA-directed RNA polymerase I subunit RPA34
+Macromolecule #15: RNA polymerase I-specific transcription initiation factor RRN6
+Macromolecule #16: RNA polymerase I-specific transcription initiation factor RRN7
+Macromolecule #17: RNA polymerase I-specific transcription initiation factor RRN11
+Macromolecule #18: RNA
+Macromolecule #19: template strand DNA
+Macromolecule #20: upstream non-template strand DNA
+Macromolecule #21: downstream non-template strand DNA
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: Quantifoil S7/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 12.0 sec. / Average electron dose: 17.2 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 5.0 µm / Calibrated defocus min: 2.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 15000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)