[English] 日本語
 Yorodumi
Yorodumi- EMDB-10489: Structure of complete, activated transcription complex Pol II-DSI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10489 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of complete, activated transcription complex Pol II-DSIF-PAF-SPT6 uncovers allosteric elongation activation by RTF1 (Map 10) | |||||||||||||||
 Map data Map data | Postprocessed map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
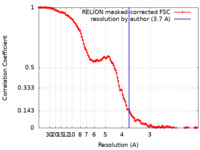
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||||||||
 Authors Authors | Vos SM / Farnung L / Cramer P | |||||||||||||||
| Funding support |  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: Structure of complete Pol II-DSIF-PAF-SPT6 transcription complex reveals RTF1 allosteric activation. Authors: Seychelle M Vos / Lucas Farnung / Andreas Linden / Henning Urlaub / Patrick Cramer /   Abstract: Transcription by RNA polymerase II (Pol II) is carried out by an elongation complex. We previously reported an activated porcine Pol II elongation complex, EC*, encompassing the human elongation ...Transcription by RNA polymerase II (Pol II) is carried out by an elongation complex. We previously reported an activated porcine Pol II elongation complex, EC*, encompassing the human elongation factors DSIF, PAF1 complex (PAF) and SPT6. Here we report the cryo-EM structure of the complete EC* that contains RTF1, a dissociable PAF subunit critical for chromatin transcription. The RTF1 Plus3 domain associates with Pol II subunit RPB12 and the phosphorylated C-terminal region of DSIF subunit SPT5. RTF1 also forms four α-helices that extend from the Plus3 domain along the Pol II protrusion and RPB10 to the polymerase funnel. The C-terminal 'fastener' helix retains PAF and is followed by a 'latch' that reaches the end of the bridge helix, a flexible element of the Pol II active site. RTF1 strongly stimulates Pol II elongation, and this requires the latch, possibly suggesting that RTF1 activates transcription allosterically by influencing Pol II translocation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10489.map.gz emd_10489.map.gz | 158.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10489-v30.xml emd-10489-v30.xml emd-10489.xml emd-10489.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10489_fsc.xml emd_10489_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10489.png emd_10489.png | 128.2 KB | ||
| Others |  emd_10489_half_map_1.map.gz emd_10489_half_map_1.map.gz emd_10489_half_map_2.map.gz emd_10489_half_map_2.map.gz | 140.3 MB 140.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10489 http://ftp.pdbj.org/pub/emdb/structures/EMD-10489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10489 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10489.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10489.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.049 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 1
| File | emd_10489_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_10489_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complete EC*
| Entire | Name: Complete EC* |
|---|---|
| Components |
|
-Supramolecule #1: Complete EC*
| Supramolecule | Name: Complete EC* / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#24 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.34 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 2 microliters applied to both sides of grid. Sample incubated on grid for 10s prior to blotting. Blotting for 8.5s.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 3 / Number real images: 13679 / Average exposure time: 10.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)