Entry Database : PDB / ID : 4ktcTitle NS3/NS4A protease with inhibitor NS4A peptide Serine protease NS3 Keywords / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Synthetic (others) Method / / Resolution : 2.3 Å Authors Zhang, H. / Ballard, J. / Vigers, G.P.A. / Brandhuber, B.J. Journal : J.Med.Chem. / Year : 2014Title : Discovery of Danoprevir (ITMN-191/R7227), a Highly Selective and Potent Inhibitor of Hepatitis C Virus (HCV) NS3/4A Protease.Authors: Jiang, Y. / Andrews, S.W. / Condroski, K.R. / Buckman, B. / Serebryany, V. / Wenglowsky, S. / Kennedy, A.L. / Madduru, M.R. / Wang, B. / Lyon, M. / Doherty, G.A. / Woodard, B.T. / Lemieux, C. ... Authors : Jiang, Y. / Andrews, S.W. / Condroski, K.R. / Buckman, B. / Serebryany, V. / Wenglowsky, S. / Kennedy, A.L. / Madduru, M.R. / Wang, B. / Lyon, M. / Doherty, G.A. / Woodard, B.T. / Lemieux, C. / Do, M.G. / Zhang, H. / Ballard, J. / Vigers, G. / Brandhuber, B.J. / Stengel, P. / Josey, J.A. / Beigelman, L. / Blatt, L. / Seiwert, S.D. History Deposition May 20, 2013 Deposition site / Processing site Revision 1.0 Aug 7, 2013 Provider / Type Revision 1.1 Mar 26, 2014 Group Revision 1.2 Sep 20, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn / struct_ref_seq_dif / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_label_asym_id / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Hepatitis C virus
Hepatitis C virus X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å
MOLECULAR REPLACEMENT / Resolution: 2.3 Å  Authors
Authors Citation
Citation Journal: J.Med.Chem. / Year: 2014
Journal: J.Med.Chem. / Year: 2014 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4ktc.cif.gz
4ktc.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4ktc.ent.gz
pdb4ktc.ent.gz PDB format
PDB format 4ktc.json.gz
4ktc.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/kt/4ktc
https://data.pdbj.org/pub/pdb/validation_reports/kt/4ktc ftp://data.pdbj.org/pub/pdb/validation_reports/kt/4ktc
ftp://data.pdbj.org/pub/pdb/validation_reports/kt/4ktc
 Links
Links Assembly
Assembly
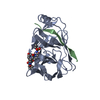

 Components
Components Hepatitis C virus (isolate Japanese) / Production host:
Hepatitis C virus (isolate Japanese) / Production host: 
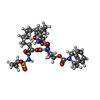
 Type: Cyclic peptide / Class: Inhibitor / Mass: 741.894 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C37H51N5O9S
Type: Cyclic peptide / Class: Inhibitor / Mass: 741.894 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C37H51N5O9S X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation ROTATING ANODE / Type: RIGAKU FR-E+ SUPERBRIGHT / Wavelength: 1.5418 Å
ROTATING ANODE / Type: RIGAKU FR-E+ SUPERBRIGHT / Wavelength: 1.5418 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj



