[English] 日本語
 Yorodumi
Yorodumi- PDB-4j22: Tankyrase 2 in complex with 3-chloro-4-(4-methyl-2-oxo-1,2-dihydr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4j22 | ||||||
|---|---|---|---|---|---|---|---|
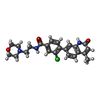
| Title | Tankyrase 2 in complex with 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide | ||||||
 Components Components | Tankyrase-2 | ||||||
 Keywords Keywords | TRANSFERASE / ribosylation | ||||||
| Function / homology |  Function and homology information Function and homology informationXAV939 stabilizes AXIN / positive regulation of telomere capping / NAD+ ADP-ribosyltransferase / protein auto-ADP-ribosylation / protein localization to chromosome, telomeric region / negative regulation of telomere maintenance via telomere lengthening / NAD+-protein-aspartate ADP-ribosyltransferase activity / protein poly-ADP-ribosylation / NAD+-protein-glutamate ADP-ribosyltransferase activity / NAD+-protein mono-ADP-ribosyltransferase activity ...XAV939 stabilizes AXIN / positive regulation of telomere capping / NAD+ ADP-ribosyltransferase / protein auto-ADP-ribosylation / protein localization to chromosome, telomeric region / negative regulation of telomere maintenance via telomere lengthening / NAD+-protein-aspartate ADP-ribosyltransferase activity / protein poly-ADP-ribosylation / NAD+-protein-glutamate ADP-ribosyltransferase activity / NAD+-protein mono-ADP-ribosyltransferase activity / pericentriolar material / Transferases; Glycosyltransferases; Pentosyltransferases / NAD+ poly-ADP-ribosyltransferase activity / positive regulation of telomere maintenance via telomerase / nucleotidyltransferase activity / TCF dependent signaling in response to WNT / Degradation of AXIN / Wnt signaling pathway / Regulation of PTEN stability and activity / protein polyubiquitination / positive regulation of canonical Wnt signaling pathway / nuclear envelope / chromosome, telomeric region / Ub-specific processing proteases / Golgi membrane / perinuclear region of cytoplasm / enzyme binding / metal ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.12 Å MOLECULAR REPLACEMENT / Resolution: 2.12 Å | ||||||
 Authors Authors | Jansson, A.E. / Larsson, E.A. / Nordlund, P.L. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2013 Journal: J.Med.Chem. / Year: 2013Title: Fragment-based ligand design of novel potent inhibitors of tankyrases. Authors: Larsson, E.A. / Jansson, A.E. / Ng, F.M. / Then, S.W. / Panicker, R. / Liu, B. / Sangthongpitag, K. / Pendharkar, V. / Tai, S.J. / Hill, J. / Dan, C. / Ho, S.Y. / Cheong, W.W. / Poulsen, A. ...Authors: Larsson, E.A. / Jansson, A.E. / Ng, F.M. / Then, S.W. / Panicker, R. / Liu, B. / Sangthongpitag, K. / Pendharkar, V. / Tai, S.J. / Hill, J. / Dan, C. / Ho, S.Y. / Cheong, W.W. / Poulsen, A. / Blanchard, S. / Lin, G.R. / Alam, J. / Keller, T.H. / Nordlund, P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4j22.cif.gz 4j22.cif.gz | 105.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4j22.ent.gz pdb4j22.ent.gz | 80.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4j22.json.gz 4j22.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/j2/4j22 https://data.pdbj.org/pub/pdb/validation_reports/j2/4j22 ftp://data.pdbj.org/pub/pdb/validation_reports/j2/4j22 ftp://data.pdbj.org/pub/pdb/validation_reports/j2/4j22 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3w51C  4iueC  4j1zC  4j21C  4j3lC  4j3mC  3kr7S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 23993.195 Da / Num. of mol.: 2 / Fragment: catalytic domai, UNP RESIDUES 952-1161 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: PARP5B, TANK2, TNKL, TNKS2 / Plasmid: pNIC-Bsa4 / Production host: Homo sapiens (human) / Gene: PARP5B, TANK2, TNKL, TNKS2 / Plasmid: pNIC-Bsa4 / Production host:  #2: Chemical | #3: Chemical | ChemComp-SO4 / #4: Chemical | #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.74 Å3/Da / Density % sol: 55.03 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 8.5 Details: 17% PEG 3350, 0.2M Ammonium sulphate, 0.1M Tris pH 8.5, vapour diffusion, hanging drop, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 277 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Australian Synchrotron Australian Synchrotron  / Beamline: MX1 / Beamline: MX1 |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Feb 10, 2012 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 2.1→116.27 Å / Num. all: 30105 / Num. obs: 30105 / Observed criterion σ(F): 0 / Observed criterion σ(I): 2 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3KR7 Resolution: 2.12→116.27 Å / Cor.coef. Fo:Fc: 0.943 / Cor.coef. Fo:Fc free: 0.912 / Occupancy max: 1 / Occupancy min: 0.5 / SU B: 4.855 / SU ML: 0.128 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.218 / ESU R Free: 0.189 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES: REFINED INDIVIDUALLY
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 77.33 Å2 / Biso mean: 25.4169 Å2 / Biso min: 2.99 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.12→116.27 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.12→2.175 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller




















 PDBj
PDBj