+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4i2d | ||||||
|---|---|---|---|---|---|---|---|
| Title | Binary complex of mouse TdT with AMPcPP | ||||||
 Components Components | DNA nucleotidylexotransferase | ||||||
 Keywords Keywords | TRANSFERASE / Terminal transferase | ||||||
| Function / homology |  Function and homology information Function and homology informationDNA nucleotidylexotransferase / DNA nucleotidylexotransferase activity / DNA modification / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / DNA metabolic process / response to ATP / euchromatin / nuclear matrix / DNA-directed DNA polymerase activity / hydrolase activity ...DNA nucleotidylexotransferase / DNA nucleotidylexotransferase activity / DNA modification / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / DNA metabolic process / response to ATP / euchromatin / nuclear matrix / DNA-directed DNA polymerase activity / hydrolase activity / DNA repair / DNA binding / nucleoplasm / metal ion binding / nucleus / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å MOLECULAR REPLACEMENT / Resolution: 2.3 Å | ||||||
 Authors Authors | Gouge, J. / Delarue, M. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2013 Journal: J.Mol.Biol. / Year: 2013Title: Structures of Intermediates along the Catalytic Cycle of Terminal Deoxynucleotidyltransferase: Dynamical Aspects of the Two-Metal Ion Mechanism. Authors: Gouge, J. / Rosario, S. / Romain, F. / Beguin, P. / Delarue, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4i2d.cif.gz 4i2d.cif.gz | 95.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4i2d.ent.gz pdb4i2d.ent.gz | 68.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4i2d.json.gz 4i2d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/i2/4i2d https://data.pdbj.org/pub/pdb/validation_reports/i2/4i2d ftp://data.pdbj.org/pub/pdb/validation_reports/i2/4i2d ftp://data.pdbj.org/pub/pdb/validation_reports/i2/4i2d | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4i27C  4i28C  4i29C  4i2aC  4i2bC  4i2cC  4i2eC  4i2fC  4i2gC  4i2hC  4i2iC  4i2jC  1jmsS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 45704.008 Da / Num. of mol.: 1 / Fragment: SEE REMARK 999 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 5 types, 266 molecules 

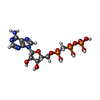






| #2: Chemical | ChemComp-MG / |
|---|---|
| #3: Chemical | ChemComp-NA / |
| #4: Chemical | ChemComp-APC / |
| #5: Chemical | ChemComp-ZN / |
| #6: Water | ChemComp-HOH / |
-Details
| Sequence details | PROTEIN FRAGMENT IS THE CATALYTIC CORE OF THE TDT-S (TDT-SMALL) ISOFORM (UNP RESIDUES 132-482, 503-530). |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.47 Å3/Da / Density % sol: 50.15 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop Details: 20-26% PEG4000, 100 mM HEPES, 200 mM ammonium acetate, pH 6.0-7.5, VAPOR DIFFUSION, HANGING DROP, temperature 293K PH range: 6.0-7.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SOLEIL SOLEIL  / Beamline: PROXIMA 1 / Wavelength: 0.915 Å / Beamline: PROXIMA 1 / Wavelength: 0.915 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Jul 18, 2012 |
| Radiation | Monochromator: channel cut / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.915 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→47 Å / Num. obs: 20328 / % possible obs: 98.3 % / Redundancy: 4.5 % / Biso Wilson estimate: 45.65 Å2 / Rmerge(I) obs: 0.1 |
| Reflection shell | Resolution: 2.3→2.42 Å / Redundancy: 4.6 % / Rmerge(I) obs: 0.695 / Mean I/σ(I) obs: 2 / Num. unique all: 2936 / % possible all: 99 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1JMS Resolution: 2.3→23.93 Å / Cor.coef. Fo:Fc: 0.8851 / Cor.coef. Fo:Fc free: 0.8756 / SU R Cruickshank DPI: 0.295 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.334 / SU Rfree Blow DPI: 0.221 / SU Rfree Cruickshank DPI: 0.214 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 49.88 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.308 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→23.93 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.3→2.42 Å / Total num. of bins used: 10
|
 Movie
Movie Controller
Controller













 PDBj
PDBj











