| Entry | Database: PDB / ID: 4hyf
|
|---|
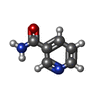
| Title | Structural basis and SAR for OD 270, a lead stage 1,2,4-triazole based specific Tankyrase1/2 inhibitor |
|---|
 Components Components | Tankyrase-2 |
|---|
 Keywords Keywords | TRANSFERASE/TRANSFERASE INHIBITOR / CATALYTIC FRAGMENT / PARP / ADP-RIBOSYLATION / ANK REPEAT / CHROMOSOMAL PROTEIN / GLYCOSYLTRANSFERASE / PHOSPHORYLATION / TELOMERE / TRANSFERASE / TRANSLOCATION / TRANSPORT / WNT-SIGNALLING / TRANSFERASE-TRANSFERASE INHIBITOR complex |
|---|
| Function / homology |  Function and homology information Function and homology information
XAV939 stabilizes AXIN / positive regulation of telomere capping / NAD+ ADP-ribosyltransferase / protein auto-ADP-ribosylation / protein localization to chromosome, telomeric region / negative regulation of telomere maintenance via telomere lengthening / NAD+-protein-aspartate ADP-ribosyltransferase activity / protein poly-ADP-ribosylation / NAD+-protein-glutamate ADP-ribosyltransferase activity / NAD+-protein mono-ADP-ribosyltransferase activity ...XAV939 stabilizes AXIN / positive regulation of telomere capping / NAD+ ADP-ribosyltransferase / protein auto-ADP-ribosylation / protein localization to chromosome, telomeric region / negative regulation of telomere maintenance via telomere lengthening / NAD+-protein-aspartate ADP-ribosyltransferase activity / protein poly-ADP-ribosylation / NAD+-protein-glutamate ADP-ribosyltransferase activity / NAD+-protein mono-ADP-ribosyltransferase activity / pericentriolar material / Transferases; Glycosyltransferases; Pentosyltransferases / NAD+ poly-ADP-ribosyltransferase activity / positive regulation of telomere maintenance via telomerase / nucleotidyltransferase activity / TCF dependent signaling in response to WNT / Degradation of AXIN / Wnt signaling pathway / Regulation of PTEN stability and activity / protein polyubiquitination / positive regulation of canonical Wnt signaling pathway / nuclear envelope / chromosome, telomeric region / Ub-specific processing proteases / Golgi membrane / perinuclear region of cytoplasm / enzyme binding / metal ion binding / nucleus / cytoplasm / cytosolSimilarity search - Function Phosphoenolpyruvate Carboxykinase; domain 3 - #10 / Phosphoenolpyruvate Carboxykinase; domain 3 / Ankyrin repeats (many copies) / Poly(ADP-ribose) polymerase catalytic domain / Poly(ADP-ribose) polymerase, catalytic domain / PARP catalytic domain profile. / SAM domain (Sterile alpha motif) / SAM domain profile. / Sterile alpha motif. / Sterile alpha motif domain ...Phosphoenolpyruvate Carboxykinase; domain 3 - #10 / Phosphoenolpyruvate Carboxykinase; domain 3 / Ankyrin repeats (many copies) / Poly(ADP-ribose) polymerase catalytic domain / Poly(ADP-ribose) polymerase, catalytic domain / PARP catalytic domain profile. / SAM domain (Sterile alpha motif) / SAM domain profile. / Sterile alpha motif. / Sterile alpha motif domain / Sterile alpha motif/pointed domain superfamily / Ankyrin repeat / Ankyrin repeats (3 copies) / Ankyrin repeat profile. / Ankyrin repeat region circular profile. / ankyrin repeats / Ankyrin repeat / Ankyrin repeat-containing domain superfamily / Alpha-Beta Complex / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.8 Å MOLECULAR REPLACEMENT / Resolution: 2.8 Å |
|---|
 Authors Authors | Perdreau, H. / Ekblad, B. / Voronkov, A. / Holsworth, D.D. / Waaler, J. / Drewes, G. / Schueler, H. / Krauss, S. / Morth, J.P. |
|---|
 Citation Citation |  Journal: J.Med.Chem. / Year: 2013 Journal: J.Med.Chem. / Year: 2013
Title: Structural Basis and SAR for G007-LK, a Lead Stage 1,2,4-Triazole Based Specific Tankyrase 1/2 Inhibitor.
Authors: Voronkov, A. / Holsworth, D.D. / Waaler, J. / Wilson, S.R. / Ekblad, B. / Perdreau-Dahl, H. / Dinh, H. / Drewes, G. / Hopf, C. / Morth, J.P. / Krauss, S. |
|---|
| History | | Deposition | Nov 13, 2012 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Mar 20, 2013 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | May 1, 2013 | Group: Database references |
|---|
| Revision 1.2 | Nov 15, 2017 | Group: Refinement description / Category: software / Item: _software.name |
|---|
| Revision 1.3 | Sep 20, 2023 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn / struct_ref_seq_dif / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_label_asym_id / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.8 Å
MOLECULAR REPLACEMENT / Resolution: 2.8 Å  Authors
Authors Citation
Citation Journal: J.Med.Chem. / Year: 2013
Journal: J.Med.Chem. / Year: 2013 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4hyf.cif.gz
4hyf.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4hyf.ent.gz
pdb4hyf.ent.gz PDB format
PDB format 4hyf.json.gz
4hyf.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/hy/4hyf
https://data.pdbj.org/pub/pdb/validation_reports/hy/4hyf ftp://data.pdbj.org/pub/pdb/validation_reports/hy/4hyf
ftp://data.pdbj.org/pub/pdb/validation_reports/hy/4hyf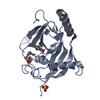
 Links
Links Assembly
Assembly



 Components
Components Homo sapiens (human) / Gene: PARP5B, TANK2, TNKL, TNKS2 / Plasmid: PNIC-BSA4 / Production host:
Homo sapiens (human) / Gene: PARP5B, TANK2, TNKL, TNKS2 / Plasmid: PNIC-BSA4 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06DA / Wavelength: 1.28243 Å
/ Beamline: X06DA / Wavelength: 1.28243 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller

















 PDBj
PDBj