[English] 日本語
 Yorodumi
Yorodumi- PDB-4gj2: Tyk2 (JH1) in complex with 2,6-dichloro-N-[2-({[(1R,2R)-2-fluoroc... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4gj2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Tyk2 (JH1) in complex with 2,6-dichloro-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide | ||||||
 Components Components | Non-receptor tyrosine-protein kinase TYK2 | ||||||
 Keywords Keywords | TRANSFERASE/TRANSFERASE INHIBITOR / Kinase / ATP Binding / TRANSFERASE-TRANSFERASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationtype III interferon-mediated signaling pathway / interleukin-10-mediated signaling pathway / interleukin-12 receptor complex / interleukin-23 receptor complex / interleukin-23-mediated signaling pathway / Interleukin-23 signaling / positive regulation of T-helper 17 type immune response / interleukin-12-mediated signaling pathway / positive regulation of NK T cell proliferation / Interleukin-12 signaling ...type III interferon-mediated signaling pathway / interleukin-10-mediated signaling pathway / interleukin-12 receptor complex / interleukin-23 receptor complex / interleukin-23-mediated signaling pathway / Interleukin-23 signaling / positive regulation of T-helper 17 type immune response / interleukin-12-mediated signaling pathway / positive regulation of NK T cell proliferation / Interleukin-12 signaling / IL-6-type cytokine receptor ligand interactions / Interleukin-27 signaling / Interleukin-35 Signalling / growth hormone receptor binding / extrinsic component of cytoplasmic side of plasma membrane / Other interleukin signaling / extrinsic component of plasma membrane / Interleukin-20 family signaling / Interleukin-6 signaling / type I interferon-mediated signaling pathway / MAPK3 (ERK1) activation / MAPK1 (ERK2) activation / Interleukin-10 signaling / positive regulation of interleukin-17 production / positive regulation of natural killer cell proliferation / Regulation of IFNA/IFNB signaling / growth hormone receptor signaling pathway via JAK-STAT / cell surface receptor signaling pathway via JAK-STAT / type II interferon-mediated signaling pathway / Signaling by CSF3 (G-CSF) / positive regulation of T cell proliferation / non-membrane spanning protein tyrosine kinase activity / positive regulation of receptor signaling pathway via JAK-STAT / non-specific protein-tyrosine kinase / cellular response to virus / Inactivation of CSF3 (G-CSF) signaling / positive regulation of protein localization to nucleus / Evasion by RSV of host interferon responses / cytoplasmic side of plasma membrane / positive regulation of type II interferon production / cytokine-mediated signaling pathway / Interferon alpha/beta signaling / Signaling by ALK fusions and activated point mutants / protein tyrosine kinase activity / Interleukin-4 and Interleukin-13 signaling / Potential therapeutics for SARS / cytoskeleton / cell differentiation / protein phosphorylation / receptor complex / cell population proliferation / intracellular signal transduction / immune response / SARS-CoV-2 activates/modulates innate and adaptive immune responses / extracellular exosome / ATP binding / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.4 Å MOLECULAR REPLACEMENT / Resolution: 2.4 Å | ||||||
 Authors Authors | Ultsch, M.H. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2013 Journal: J.Med.Chem. / Year: 2013Title: Lead Optimization of a 4-Aminopyridine Benzamide Scaffold To Identify Potent, Selective, and Orally Bioavailable TYK2 Inhibitors. Authors: Liang, J. / van Abbema, A. / Balazs, M. / Barrett, K. / Berezhkovsky, L. / Blair, W. / Chang, C. / Delarosa, D. / Devoss, J. / Driscoll, J. / Eigenbrot, C. / Ghilardi, N. / Gibbons, P. / ...Authors: Liang, J. / van Abbema, A. / Balazs, M. / Barrett, K. / Berezhkovsky, L. / Blair, W. / Chang, C. / Delarosa, D. / Devoss, J. / Driscoll, J. / Eigenbrot, C. / Ghilardi, N. / Gibbons, P. / Halladay, J. / Johnson, A. / Kohli, P.B. / Lai, Y. / Liu, Y. / Lyssikatos, J. / Mantik, P. / Menghrajani, K. / Murray, J. / Peng, I. / Sambrone, A. / Shia, S. / Shin, Y. / Smith, J. / Sohn, S. / Tsui, V. / Ultsch, M. / Wu, L.C. / Xiao, Y. / Yang, W. / Young, J. / Zhang, B. / Zhu, B.Y. / Magnuson, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4gj2.cif.gz 4gj2.cif.gz | 134.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4gj2.ent.gz pdb4gj2.ent.gz | 103.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4gj2.json.gz 4gj2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gj/4gj2 https://data.pdbj.org/pub/pdb/validation_reports/gj/4gj2 ftp://data.pdbj.org/pub/pdb/validation_reports/gj/4gj2 ftp://data.pdbj.org/pub/pdb/validation_reports/gj/4gj2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4giiC  4gj3C  3nz0S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 34748.734 Da / Num. of mol.: 1 / Fragment: Kinase domain, UNP residues 696-1022 / Mutation: C936A,Q969A,E971A,K972A,D1023N,C1142A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: Tyk2 gene / Production host: Homo sapiens (human) / Gene: Tyk2 gene / Production host:  References: UniProt: P29597, non-specific protein-tyrosine kinase |
|---|---|
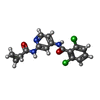
| #2: Chemical | ChemComp-0XH / |
| #3: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.04 Å3/Da / Density % sol: 39.77 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 20-25%(w/v) PEG3350 and 0.2M Mg sulfate, 0.1M MES pH6.5, VAPOR DIFFUSION, HANGING DROP, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 173 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL11-1 / Wavelength: 0.97 Å / Beamline: BL11-1 / Wavelength: 0.97 Å |
| Detector | Type: PSI PILATUS 6M / Detector: PIXEL / Date: Apr 11, 2012 |
| Radiation | Monochromator: Side scattering bent cube-root I-beam single crystal; asymmetric cut 4.965 degs Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97 Å / Relative weight: 1 |
| Reflection | Resolution: 2.4→37.04 Å / Num. obs: 11482 / % possible obs: 98.8 % / Observed criterion σ(I): 2 / Redundancy: 4 % / Biso Wilson estimate: 47.22 Å2 / Rmerge(I) obs: 0.09 / Net I/σ(I): 13 |
| Reflection shell | Resolution: 2.4→2.53 Å / Redundancy: 4.1 % / Rmerge(I) obs: 0.489 / Mean I/σ(I) obs: 2.7 / Num. unique all: 1617 / % possible all: 98.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3NZ0 Resolution: 2.4→37.04 Å / Cor.coef. Fo:Fc: 0.9339 / Cor.coef. Fo:Fc free: 0.8881 / SU R Cruickshank DPI: 0.576 / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 2 / Stereochemistry target values: Engh & Huber
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 42.82 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.313 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.4→37.04 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.4→2.63 Å / Total num. of bins used: 6
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj