[English] 日本語
 Yorodumi
Yorodumi- PDB-4ejr: Crystal structure of major capsid protein S domain from rabbit he... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4ejr | ||||||
|---|---|---|---|---|---|---|---|
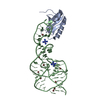
| Title | Crystal structure of major capsid protein S domain from rabbit hemorrhagic disease virus | ||||||
 Components Components | Major capsid protein VP60 | ||||||
 Keywords Keywords | VIRAL PROTEIN / capsid protein | ||||||
| Function / homology |  Function and homology information Function and homology informationvirion component / ribonucleoside triphosphate phosphatase activity / viral capsid / host cell cytoplasm / RNA helicase activity / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / proteolysis ...virion component / ribonucleoside triphosphate phosphatase activity / viral capsid / host cell cytoplasm / RNA helicase activity / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / proteolysis / RNA binding / ATP binding Similarity search - Function | ||||||
| Biological species |  Rabbit hemorrhagic disease virus Rabbit hemorrhagic disease virus | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Xu, F. / Ma, J. / Zhang, K. / Wang, X. / Sun, F. | ||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2013 Journal: PLoS Pathog / Year: 2013Title: Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography. Authors: Xue Wang / Fengting Xu / Jiasen Liu / Bingquan Gao / Yanxin Liu / Yujia Zhai / Jun Ma / Kai Zhang / Timothy S Baker / Klaus Schulten / Dong Zheng / Hai Pang / Fei Sun /  Abstract: Rabbit hemorrhagic disease, first described in China in 1984, causes hemorrhagic necrosis of the liver. Its etiological agent, rabbit hemorrhagic disease virus (RHDV), belongs to the Lagovirus genus ...Rabbit hemorrhagic disease, first described in China in 1984, causes hemorrhagic necrosis of the liver. Its etiological agent, rabbit hemorrhagic disease virus (RHDV), belongs to the Lagovirus genus in the family Caliciviridae. The detailed molecular structure of any lagovirus capsid has yet to be determined. Here, we report a cryo-electron microscopic (cryoEM) reconstruction of wild-type RHDV at 6.5 Å resolution and the crystal structures of the shell (S) and protruding (P) domains of its major capsid protein, VP60, each at 2.0 Å resolution. From these data we built a complete atomic model of the RHDV capsid. VP60 has a conserved S domain and a specific P2 sub-domain that differs from those found in other caliciviruses. As seen in the shell portion of the RHDV cryoEM map, which was resolved to ~5.5 Å, the N-terminal arm domain of VP60 folds back onto its cognate S domain. Sequence alignments of VP60 from six groups of RHDV isolates revealed seven regions of high variation that could be mapped onto the surface of the P2 sub-domain and suggested three putative pockets might be responsible for binding to histo-blood group antigens. A flexible loop in one of these regions was shown to interact with rabbit tissue cells and contains an important epitope for anti-RHDV antibody production. Our study provides a reliable, pseudo-atomic model of a Lagovirus and suggests a new candidate for an efficient vaccine that can be used to protect rabbits from RHDV infection. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4ejr.cif.gz 4ejr.cif.gz | 79.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4ejr.ent.gz pdb4ejr.ent.gz | 57.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4ejr.json.gz 4ejr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ej/4ejr https://data.pdbj.org/pub/pdb/validation_reports/ej/4ejr ftp://data.pdbj.org/pub/pdb/validation_reports/ej/4ejr ftp://data.pdbj.org/pub/pdb/validation_reports/ej/4ejr | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5410C  3j1pC  4egtC  2gh8S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 27208.283 Da / Num. of mol.: 2 / Fragment: S (inner shell) domain (UNP residues 1-230) Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Rabbit hemorrhagic disease virus / Strain: JX/CHA/97 / Gene: VP60 / Plasmid: pEXS-DH / Production host: Rabbit hemorrhagic disease virus / Strain: JX/CHA/97 / Gene: VP60 / Plasmid: pEXS-DH / Production host:  #2: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.79 Å3/Da / Density % sol: 31.18 % |
|---|---|
| Crystal grow | Temperature: 289 K / Method: vapor diffusion / pH: 7 Details: 0.2 M magnesium chloride, 0.1 M HEPES sodium, 30% PEG400, pH 7.0, VAPOR DIFFUSION, temperature 289K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Photon Factory Photon Factory  / Beamline: BL-17A / Wavelength: 0.98 Å / Beamline: BL-17A / Wavelength: 0.98 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Jun 22, 2010 |
| Radiation | Monochromator: double crystal Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.98 Å / Relative weight: 1 |
| Reflection | Resolution: 2→50 Å / Num. all: 24872 / Num. obs: 24815 / % possible obs: 99.77 % / Observed criterion σ(F): 0 / Observed criterion σ(I): -2 / Redundancy: 7.4 % / Rmerge(I) obs: 0.086 / Net I/σ(I): 25.7 |
| Reflection shell | Resolution: 2→2.07 Å / Redundancy: 6.8 % / Rmerge(I) obs: 0.404 / Mean I/σ(I) obs: 3.97 / % possible all: 99.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2GH8 Resolution: 2→49.35 Å / Cor.coef. Fo:Fc: 0.947 / Cor.coef. Fo:Fc free: 0.931 / Cross valid method: THROUGHOUT / ESU R: 0.173 / ESU R Free: 0.159 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 23.968 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→49.35 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2→2.053 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller












 PDBj
PDBj


