[English] 日本語
 Yorodumi
Yorodumi- PDB-4cdx: Crystal structure of human Enterovirus 71 in complex with the unc... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4cdx | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP12 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | VIRUS / HAND-FOOT-AND-MOUTH DISEASE / ENTEROVIRUS UNCOATING / INHIBITOR | ||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / ribonucleoside triphosphate phosphatase activity / nucleoside-triphosphate phosphatase / channel activity ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / ribonucleoside triphosphate phosphatase activity / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | ||||||
| Biological species |   ENTEROVIRUS A71 ENTEROVIRUS A71 | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / OTHER / Resolution: 2.8 Å SYNCHROTRON / OTHER / Resolution: 2.8 Å | ||||||
 Authors Authors | De Colibus, L. / Wang, X. / Spyrou, J.A.B. / Kelly, J. / Ren, J. / Grimes, J. / Puerstinger, G. / Stonehouse, N. / Walter, T.S. / Hu, Z. ...De Colibus, L. / Wang, X. / Spyrou, J.A.B. / Kelly, J. / Ren, J. / Grimes, J. / Puerstinger, G. / Stonehouse, N. / Walter, T.S. / Hu, Z. / Wang, J. / Li, X. / Peng, W. / Rowlands, D. / Fry, E.E. / Rao, Z. / Stuart, D.I. | ||||||
 Citation Citation |  Journal: Nat.Struct.Mol.Biol. / Year: 2014 Journal: Nat.Struct.Mol.Biol. / Year: 2014Title: More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules. Authors: De Colibus, L. / Wang, X. / Spyrou, J.A.B. / Kelly, J. / Ren, J. / Grimes, J. / Puerstinger, G. / Stonehouse, N. / Walter, T.S. / Hu, Z. / Wang, J. / Li, X. / Peng, W. / Rowlands, D.J. / ...Authors: De Colibus, L. / Wang, X. / Spyrou, J.A.B. / Kelly, J. / Ren, J. / Grimes, J. / Puerstinger, G. / Stonehouse, N. / Walter, T.S. / Hu, Z. / Wang, J. / Li, X. / Peng, W. / Rowlands, D.J. / Fry, E.E. / Rao, Z. / Stuart, D.I. #1:  Journal: Nat.Struct.Mol.Biol. / Year: 2012 Journal: Nat.Struct.Mol.Biol. / Year: 2012Title: A Sensor-Adaptor Mechanism for Enterovirus Uncoating from Structures of Ev71. Authors: Wang, X. / Peng, W. / Ren, J. / Hu, Z. / Xu, J. / Lou, Z. / Li, X. / Yin, W. / Shen, X. / Porta, C. / Walter, T.S. / Evans, G. / Axford, D. / Owen, R. / Rowlands, D.J. / Wang, J. / Stuart, D. ...Authors: Wang, X. / Peng, W. / Ren, J. / Hu, Z. / Xu, J. / Lou, Z. / Li, X. / Yin, W. / Shen, X. / Porta, C. / Walter, T.S. / Evans, G. / Axford, D. / Owen, R. / Rowlands, D.J. / Wang, J. / Stuart, D.I. / Fry, E.E. / Rao, Z. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4cdx.cif.gz 4cdx.cif.gz | 180.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4cdx.ent.gz pdb4cdx.ent.gz | 142.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4cdx.json.gz 4cdx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cd/4cdx https://data.pdbj.org/pub/pdb/validation_reports/cd/4cdx ftp://data.pdbj.org/pub/pdb/validation_reports/cd/4cdx ftp://data.pdbj.org/pub/pdb/validation_reports/cd/4cdx | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | x 60
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | x 5
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | x 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: (Schoenflies symbol: I (icosahedral)) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
-Protein , 4 types, 4 molecules ABCD
| #1: Protein | Mass: 32699.840 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   ENTEROVIRUS A71 / Cell line (production host): VERO CELLS / Production host: ENTEROVIRUS A71 / Cell line (production host): VERO CELLS / Production host:  CHLOROCEBUS AETHIOPS (grivet) / References: UniProt: B2ZUN0 CHLOROCEBUS AETHIOPS (grivet) / References: UniProt: B2ZUN0 |
|---|---|
| #2: Protein | Mass: 27740.160 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   ENTEROVIRUS A71 / Cell line (production host): VERO CELLS / Production host: ENTEROVIRUS A71 / Cell line (production host): VERO CELLS / Production host:  CHLOROCEBUS AETHIOPS (grivet) / References: UniProt: B2ZUN0 CHLOROCEBUS AETHIOPS (grivet) / References: UniProt: B2ZUN0 |
| #3: Protein | Mass: 26468.225 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   ENTEROVIRUS A71 / Cell line (production host): VERO CELLS / Production host: ENTEROVIRUS A71 / Cell line (production host): VERO CELLS / Production host:  CHLOROCEBUS AETHIOPS (grivet) / References: UniProt: B2ZUN0 CHLOROCEBUS AETHIOPS (grivet) / References: UniProt: B2ZUN0 |
| #4: Protein | Mass: 7501.162 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   ENTEROVIRUS A71 / Cell line (production host): VERO CELLS / Production host: ENTEROVIRUS A71 / Cell line (production host): VERO CELLS / Production host:  CHLOROCEBUS AETHIOPS (grivet) / References: UniProt: B2ZUN0 CHLOROCEBUS AETHIOPS (grivet) / References: UniProt: B2ZUN0 |
-Non-polymers , 3 types, 113 molecules 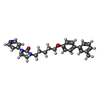




| #5: Chemical | ChemComp-JF0 / | ||
|---|---|---|---|
| #6: Chemical | | #7: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 13 X-RAY DIFFRACTION / Number of used crystals: 13 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density % sol: 75 % / Description: NONE |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 8.5 Details: 30% PEG400, 0.2 M TRI-SODIUM CITRATE, 0.1 M TRIS.HCL PH 8.5, MIXED WITH VIRUS AND EQUILIBRATED AGAINST SALT RESERVOIR, VAPOR DIFFUSION, SITTING DROP, TEMPERATURE 293K |
-Data collection
| Diffraction | Mean temperature: 293 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I24 / Wavelength: 0.9686 / Beamline: I24 / Wavelength: 0.9686 |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Oct 22, 2012 / Details: MIRRORS |
| Radiation | Monochromator: DCM / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9686 Å / Relative weight: 1 |
| Reflection | Resolution: 2.8→50 Å / Num. obs: 583785 / % possible obs: 67.4 % / Observed criterion σ(I): -3 / Redundancy: 1.9 % / Biso Wilson estimate: 27.3 Å2 / Rmerge(I) obs: 0.54 / Net I/σ(I): 1.83 |
| Reflection shell | Resolution: 2.8→2.9 Å / Redundancy: 1.8 % / Mean I/σ(I) obs: 0.57 / % possible all: 66.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: OTHER Starting model: NONE Resolution: 2.8→49.98 Å / Rfactor Rfree error: 0.002 / Data cutoff high absF: 69097870.47 / Data cutoff low absF: 0 / Isotropic thermal model: GROUP / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: BULK SOLVENT MODEL USED
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 11.771 Å2 / ksol: 0.32 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 21.3 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.8→49.98 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | NCS model details: CONSTR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.8→2.9 Å / Rfactor Rfree error: 0.007 / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller



























 PDBj
PDBj



