[English] 日本語
 Yorodumi
Yorodumi- PDB-4bog: The structure and super-organization of acetylcholine receptor-ra... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4bog | ||||||
|---|---|---|---|---|---|---|---|
| Title | The structure and super-organization of acetylcholine receptor-rapsyn complexes | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSPORT PROTEIN / CLUSTERING / SYNAPSE / NEUROMUSCULAR JUNCTION / NICOTINIC / RAPSYN / 43K / ELECTRIC ORGAN | ||||||
| Function / homology |  Function and homology information Function and homology informationacetylcholine-gated channel complex / acetylcholine-gated monoatomic cation-selective channel activity / acetylcholine receptor signaling pathway / transmembrane signaling receptor activity / postsynaptic membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | ELECTRON MICROSCOPY / electron tomography / cryo EM / Resolution: 50 Å | ||||||
 Authors Authors | Zuber, B. / Unwin, N. | ||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2013 Journal: Proc Natl Acad Sci U S A / Year: 2013Title: Structure and superorganization of acetylcholine receptor-rapsyn complexes. Authors: Benoît Zuber / Nigel Unwin /  Abstract: The scaffolding protein at the neuromuscular junction, rapsyn, enables clustering of nicotinic acetylcholine receptors in high concentration and is critical for muscle function. Patients with ...The scaffolding protein at the neuromuscular junction, rapsyn, enables clustering of nicotinic acetylcholine receptors in high concentration and is critical for muscle function. Patients with insufficient receptor clustering suffer from muscle weakness. However, the detailed organization of the receptor-rapsyn network is poorly understood: it is unclear whether rapsyn first forms a wide meshwork to which receptors can subsequently dock or whether it only forms short bridges linking receptors together to make a large cluster. Furthermore, the number of rapsyn-binding sites per receptor (a heteropentamer) has been controversial. Here, we show by cryoelectron tomography and subtomogram averaging of Torpedo postsynaptic membrane that receptors are connected by up to three rapsyn bridges, the minimum number required to form a 2D network. Half of the receptors belong to rapsyn-connected groups comprising between two and fourteen receptors. Our results provide a structural basis for explaining the stability and low diffusion of receptors within clusters. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4bog.cif.gz 4bog.cif.gz | 1.9 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4bog.ent.gz pdb4bog.ent.gz | 1.5 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4bog.json.gz 4bog.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bo/4bog https://data.pdbj.org/pub/pdb/validation_reports/bo/4bog ftp://data.pdbj.org/pub/pdb/validation_reports/bo/4bog ftp://data.pdbj.org/pub/pdb/validation_reports/bo/4bog | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 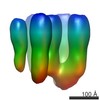 2376MC  2377C  2378C 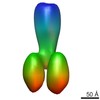 2381C  2382C 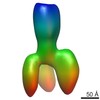 2383C  4boiC  4bonC  4booC  4borC  4botC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 56123.594 Da / Num. of mol.: 6 / Source method: isolated from a natural source / Source: (natural)  #2: Protein | Mass: 60017.684 Da / Num. of mol.: 6 / Source method: isolated from a natural source / Source: (natural)  #3: Protein | Mass: 52845.523 Da / Num. of mol.: 12 / Source method: isolated from a natural source / Source: (natural)  #4: Protein | Mass: 58118.012 Da / Num. of mol.: 6 / Source method: isolated from a natural source / Source: (natural)  Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: electron tomography |
- Sample preparation
Sample preparation
| Component | Name: ACETYLCHOLINE RECEPTOR- RAPSYN COMPLEX / Type: COMPLEX |
|---|---|
| Buffer solution | Name: 400 MM NACL, 20 MM PHOSPHATE BUFFER, LEUPEPTIN 0.3 MG/L, PEPSTATIN 1 MG/L pH: 7.4 Details: 400 MM NACL, 20 MM PHOSPHATE BUFFER, LEUPEPTIN 0.3 MG/L, PEPSTATIN 1 MG/L |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Details: HOLEY CARBON |
| Vitrification | Instrument: HOMEMADE PLUNGER / Cryogen name: ETHANE / Details: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TECNAI F30 / Date: Jul 25, 2008 |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 69000 X / Calibrated magnification: 80213 X / Nominal defocus max: 6000 nm / Nominal defocus min: 3000 nm / Cs: 2 mm |
| Specimen holder | Temperature: 85 K / Tilt angle max: 70 ° / Tilt angle min: -70 ° |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) |
| Image scans | Num. digital images: 142 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||
| 3D reconstruction | Method: MAXIMUM LIKELIHOOD SUBTOMOGRAM AVERAGING / Resolution: 50 Å / Num. of particles: 3564 / Nominal pixel size: 7.48 Å / Actual pixel size: 7.48 Å / Magnification calibration: CATALASE CRYSTAL Details: SUBMISSION BASED ON EXPERIMENTAL DATA FROM EMDB EMD-2376. (DEPOSITION ID: 11658). Symmetry type: POINT | ||||||||||||||||||||||||
| Atomic model building | Protocol: OTHER / Space: REAL Details: METHOD--LOCAL CORRELATION REFINEMENT PROTOCOL--ELECTRON MICROSCOPY | ||||||||||||||||||||||||
| Atomic model building | PDB-ID: 2BG9 Accession code: 2BG9 / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||
| Refinement | Highest resolution: 50 Å | ||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 50 Å
|
 Movie
Movie Controller
Controller



 PDBj
PDBj


