[English] 日本語
 Yorodumi
Yorodumi- PDB-4a1o: Crystal structure of Mycobacterium tuberculosis PurH complexed wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4a1o | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Mycobacterium tuberculosis PurH complexed with AICAR and a novel nucleotide CFAIR, at 2.48 A resolution. | ||||||
 Components Components | BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH | ||||||
 Keywords Keywords | TRANSFERASE-HYDROLASE | ||||||
| Function / homology |  Function and homology information Function and homology informationphosphoribosylaminoimidazolecarboxamide formyltransferase / phosphoribosylaminoimidazolecarboxamide formyltransferase activity / IMP cyclohydrolase / IMP cyclohydrolase activity / 'de novo' IMP biosynthetic process / plasma membrane / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.48 Å MOLECULAR REPLACEMENT / Resolution: 2.48 Å | ||||||
 Authors Authors | Le Nours, J. / Bulloch, E.M.M. / Zhang, Z. / Greenwood, D.R. / Middleditch, M.J. / Dickson, J.M.J. / Baker, E.N. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2011 Journal: J.Biol.Chem. / Year: 2011Title: Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide. Authors: Le Nours, J. / Bulloch, E.M.M. / Zhang, Z. / Greenwood, D.R. / Middleditch, M.J. / Dickson, J.M.J. / Baker, E.N. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4a1o.cif.gz 4a1o.cif.gz | 396 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4a1o.ent.gz pdb4a1o.ent.gz | 324.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4a1o.json.gz 4a1o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4a1o_validation.pdf.gz 4a1o_validation.pdf.gz | 1.4 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4a1o_full_validation.pdf.gz 4a1o_full_validation.pdf.gz | 1.4 MB | Display | |
| Data in XML |  4a1o_validation.xml.gz 4a1o_validation.xml.gz | 39.7 KB | Display | |
| Data in CIF |  4a1o_validation.cif.gz 4a1o_validation.cif.gz | 55.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/a1/4a1o https://data.pdbj.org/pub/pdb/validation_reports/a1/4a1o ftp://data.pdbj.org/pub/pdb/validation_reports/a1/4a1o ftp://data.pdbj.org/pub/pdb/validation_reports/a1/4a1o | HTTPS FTP |
-Related structure data
| Related structure data |  3zzmSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.9684, -0.2122, 0.1307), Vector: |
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 55084.949 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: P67541, UniProt: P9WHM7*PLUS, phosphoribosylaminoimidazolecarboxamide formyltransferase, IMP cyclohydrolase |
|---|
-Non-polymers , 5 types, 279 molecules 


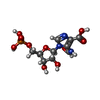





| #2: Chemical | | #3: Chemical | #4: Chemical | #5: Chemical | ChemComp-JLN / | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.3 Å3/Da / Density % sol: 46.5 % / Description: NONE |
|---|---|
| Crystal grow | pH: 6.5 Details: 24-34% PEG 8000, 0.2 M SODIUM ACETATE, 0.1 M SODIUM CACODYLATE PH 6.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-2 / Wavelength: 0.979 / Beamline: BL9-2 / Wavelength: 0.979 |
| Detector | Type: MARMOSAIC 325 mm CCD / Detector: CCD / Date: Jul 29, 2007 |
| Radiation | Monochromator: DOUBLE CRYSTAL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.979 Å / Relative weight: 1 |
| Reflection | Resolution: 2.48→50 Å / Num. obs: 38276 / % possible obs: 98.8 % / Observed criterion σ(I): 2 / Redundancy: 10.5 % / Biso Wilson estimate: 59.45 Å2 / Rmerge(I) obs: 0.12 / Net I/σ(I): 20.8 |
| Reflection shell | Resolution: 2.2→2.58 Å / Redundancy: 7 % / Rmerge(I) obs: 0.39 / Mean I/σ(I) obs: 4.7 / % possible all: 88.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 3ZZM Resolution: 2.48→43.84 Å / Cor.coef. Fo:Fc: 0.95 / Cor.coef. Fo:Fc free: 0.9241 / Cross valid method: THROUGHOUT / σ(F): 0 Details: REFINEMENT NOTE 1: IDEAL-DIST CONTACT TERM CONTACT SETUP. RESIDUE TYPES WITHOUT CCP4 ATOM TYPE IN LIBRARY=K AMZ JLN. NUMBER OF ATOMS WITH PROPER CCP4 ATOM TYPE= 7962. NUMBER WITH APPROX ...Details: REFINEMENT NOTE 1: IDEAL-DIST CONTACT TERM CONTACT SETUP. RESIDUE TYPES WITHOUT CCP4 ATOM TYPE IN LIBRARY=K AMZ JLN. NUMBER OF ATOMS WITH PROPER CCP4 ATOM TYPE= 7962. NUMBER WITH APPROX DEFAULT CCP4 ATOM TYPE=68. NUMBER RESTRAINT LSSR (-AUTONCS).
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 51.97 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.48→43.84 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.48→2.55 Å / Total num. of bins used: 19
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
|
 Movie
Movie Controller
Controller












 PDBj
PDBj




