+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 484d | ||||||
|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF HIV-1 REV PEPTIDE-RNA APTAMER COMPLEX | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA BINDING PROTEIN/RNA / HIV-1 REV PEPTIDE / RNA APTAMER / BOUND PEPTIDE SECONDARY STRUCTURE / PEPTIDE- BINDING RNA / TERTIARY ARCHITECTURES / ADAPTIVE-BINDING / RNA BINDING PROTEIN-RNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationhost cell nucleolus / mRNA transport / host cell cytoplasm / DNA-binding transcription factor activity / RNA binding Similarity search - Function | ||||||
| Method | SOLUTION NMR / DIHEDRAL ANGLE MD | ||||||
 Authors Authors | Ye, X. / Gorin, A.A. / Frederick, R. / Hu, W. / Majumdar, A. / Xu, W. / Mclendon, G. / Ellington, A. / Patel, D.J. | ||||||
 Citation Citation |  Journal: Chem.Biol. / Year: 1999 Journal: Chem.Biol. / Year: 1999Title: RNA architecture dictates the conformations of a bound peptide. Authors: Ye, X. / Gorin, A. / Frederick, R. / Hu, W. / Majumdar, A. / Xu, W. / McLendon, G. / Ellington, A. / Patel, D.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  484d.cif.gz 484d.cif.gz | 530 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb484d.ent.gz pdb484d.ent.gz | 438.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  484d.json.gz 484d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  484d_validation.pdf.gz 484d_validation.pdf.gz | 382.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  484d_full_validation.pdf.gz 484d_full_validation.pdf.gz | 626.9 KB | Display | |
| Data in XML |  484d_validation.xml.gz 484d_validation.xml.gz | 33.3 KB | Display | |
| Data in CIF |  484d_validation.cif.gz 484d_validation.cif.gz | 53.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/84/484d https://data.pdbj.org/pub/pdb/validation_reports/84/484d ftp://data.pdbj.org/pub/pdb/validation_reports/84/484d ftp://data.pdbj.org/pub/pdb/validation_reports/84/484d | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 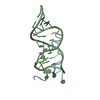
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 8698.176 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: Protein/peptide | Mass: 2447.819 Da / Num. of mol.: 1 / Fragment: RESIDUES 34-50 / Source method: obtained synthetically Details: SEQUENCE FROM REV PROTEIN OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 References: UniProt: Q9IQN3, UniProt: Q79994*PLUS |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR experiment | Type: SEE BELOW |
| NMR details | Text: 2D NOESY (H2O AND D2O), 2D DQ-COSY, 2D TOCSY, 2D 1H-15N HSQC (H2O), 2D 1H-13C HSQC(D2O), 2D 15N-EDITED HMQC-NOESY (H2O), 3D HCCH-COSY (D2O), 3D HCCH-TOCSY ( D2O) AND 3D 13C-EDITED NOESY-HMQC (D2O) |
- Sample preparation
Sample preparation
| Details | Contents: H2O, D2O |
|---|---|
| Sample conditions | Ionic strength: 10 mM SODIUM PHOSPHATE / pH: 6.2 / Pressure: 1 atm / Temperature: 297 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: DIHEDRAL ANGLE MD / Software ordinal: 1 Details: REFINEMENT DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE. | ||||||||||||
| NMR ensemble | Conformer selection criteria: LEAST RESTRAINT VIOLATION / Conformers calculated total number: 60 / Conformers submitted total number: 22 |
 Movie
Movie Controller
Controller












 PDBj
PDBj





























 X-PLOR
X-PLOR