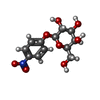
| Entry | Database: PDB / ID: 3zyf
|
|---|
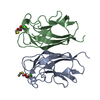
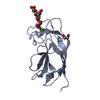
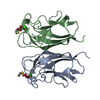
| Title | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH NPG AT 1.9 A RESOLUTION |
|---|
 Components Components | PA-I GALACTOPHILIC LECTIN |
|---|
 Keywords Keywords | SUGAR BINDING PROTEIN / ADHESIN / GLYCOSPHINGOLIPID-ANTIGEN / GALACTOSE-SPECIFIC / GALACTOSIDES |
|---|
| Function / homology |  Function and homology information Function and homology information
PA-IL-like / PA-IL-like protein / Galactose-binding lectin / Galactose-binding-like domain superfamily / Jelly Rolls / Sandwich / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |   PSEUDOMONAS AERUGINOSA (bacteria) PSEUDOMONAS AERUGINOSA (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.944 Å MOLECULAR REPLACEMENT / Resolution: 1.944 Å |
|---|
 Authors Authors | Kadam, R.U. / Bergmann, M. / Hurley, M. / Garg, D. / Cacciarini, M. / Swiderska, M.A. / Nativi, C. / Sattler, M. / Smyth, A.R. / Williams, P. ...Kadam, R.U. / Bergmann, M. / Hurley, M. / Garg, D. / Cacciarini, M. / Swiderska, M.A. / Nativi, C. / Sattler, M. / Smyth, A.R. / Williams, P. / Camara, M. / Stocker, A. / Darbre, T. / Reymond, J.-L. |
|---|
 Citation Citation |  Journal: Angew.Chem.Int.Ed.Engl. / Year: 2011 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2011
Title: A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms.
Authors: Kadam, R.U. / Bergmann, M. / Hurley, M. / Garg, D. / Cacciarini, M. / Swiderska, M.A. / Nativi, C. / Sattler, M. / Smyth, A.R. / Williams, P. / Camara, M. / Stocker, A. / Darbre, T. / Reymond, J.-L. |
|---|
| History | | Deposition | Aug 22, 2011 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Sep 21, 2011 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Apr 25, 2012 | Group: Other |
|---|
| Revision 2.0 | Jul 29, 2020 | Group: Atomic model / Data collection ...Atomic model / Data collection / Derived calculations / Other / Structure summary
Category: atom_site / chem_comp ...atom_site / chem_comp / entity / pdbx_chem_comp_identifier / pdbx_database_status / pdbx_entity_nonpoly / pdbx_struct_conn_angle / struct_conn / struct_site / struct_site_gen
Item: _atom_site.auth_atom_id / _atom_site.label_atom_id ..._atom_site.auth_atom_id / _atom_site.label_atom_id / _chem_comp.mon_nstd_flag / _chem_comp.name / _entity.pdbx_description / _pdbx_database_status.status_code_sf / _pdbx_entity_nonpoly.name / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id
Description: Carbohydrate remediation / Provider: repository / Type: Remediation |
|---|
| Revision 2.1 | Dec 20, 2023 | Group: Data collection / Database references ...Data collection / Database references / Refinement description / Structure summary
Category: chem_comp / chem_comp_atom ...chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.944 Å
MOLECULAR REPLACEMENT / Resolution: 1.944 Å  Authors
Authors Citation
Citation Journal: Angew.Chem.Int.Ed.Engl. / Year: 2011
Journal: Angew.Chem.Int.Ed.Engl. / Year: 2011 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3zyf.cif.gz
3zyf.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3zyf.ent.gz
pdb3zyf.ent.gz PDB format
PDB format 3zyf.json.gz
3zyf.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/zy/3zyf
https://data.pdbj.org/pub/pdb/validation_reports/zy/3zyf ftp://data.pdbj.org/pub/pdb/validation_reports/zy/3zyf
ftp://data.pdbj.org/pub/pdb/validation_reports/zy/3zyf

 Links
Links Assembly
Assembly


 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06SA / Wavelength: 1
/ Beamline: X06SA / Wavelength: 1  Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller














 PDBj
PDBj