[English] 日本語
 Yorodumi
Yorodumi- PDB-3v4o: Human MALT1 (caspase domain) in complex with an irreversible pept... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3v4o | ||||||
|---|---|---|---|---|---|---|---|
| Title | Human MALT1 (caspase domain) in complex with an irreversible peptidic inhibitor | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE/INHIBITOR / Caspase / Hydrolase / TRAF6 / BCL10 / Cytosolic / HYDROLASE-INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationpolkadots / B-1 B cell differentiation / positive regulation of T-helper 17 cell differentiation / CBM complex / regulation of T cell receptor signaling pathway / response to fungus / CLEC7A/inflammasome pathway / nuclear export / small molecule binding / B cell activation ...polkadots / B-1 B cell differentiation / positive regulation of T-helper 17 cell differentiation / CBM complex / regulation of T cell receptor signaling pathway / response to fungus / CLEC7A/inflammasome pathway / nuclear export / small molecule binding / B cell activation / endopeptidase activator activity / T cell proliferation / positive regulation of interleukin-2 production / : / lipopolysaccharide-mediated signaling pathway / positive regulation of protein ubiquitination / positive regulation of interleukin-1 beta production / Activation of NF-kappaB in B cells / defense response / positive regulation of T cell cytokine production / CLEC7A (Dectin-1) signaling / FCERI mediated NF-kB activation / fibrillar center / ubiquitin-protein transferase activity / Downstream TCR signaling / peptidase activity / T cell receptor signaling pathway / protease binding / endopeptidase activity / regulation of apoptotic process / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / positive regulation of canonical NF-kappaB signal transduction / innate immune response / cysteine-type endopeptidase activity / negative regulation of apoptotic process / perinuclear region of cytoplasm / protein-containing complex / proteolysis / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | ||||||
 Authors Authors | Renatus, M. / Wiesmann, C. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2012 Journal: J.Mol.Biol. / Year: 2012Title: Structural Determinants of MALT1 Protease Activity. Authors: Wiesmann, C. / Leder, L. / Blank, J. / Bernardi, A. / Melkko, S. / Decock, A. / D'Arcy, A. / Villard, F. / Erbel, P. / Hughes, N. / Freuler, F. / Nikolay, R. / Alves, J. / Bornancin, F. / Renatus, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3v4o.cif.gz 3v4o.cif.gz | 64.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3v4o.ent.gz pdb3v4o.ent.gz | 47.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3v4o.json.gz 3v4o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3v4o_validation.pdf.gz 3v4o_validation.pdf.gz | 432.6 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3v4o_full_validation.pdf.gz 3v4o_full_validation.pdf.gz | 434.9 KB | Display | |
| Data in XML |  3v4o_validation.xml.gz 3v4o_validation.xml.gz | 12.6 KB | Display | |
| Data in CIF |  3v4o_validation.cif.gz 3v4o_validation.cif.gz | 17.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v4/3v4o https://data.pdbj.org/pub/pdb/validation_reports/v4/3v4o ftp://data.pdbj.org/pub/pdb/validation_reports/v4/3v4o ftp://data.pdbj.org/pub/pdb/validation_reports/v4/3v4o | HTTPS FTP |
-Related structure data
| Related structure data |  3v4lC  3v55C  2pyoS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 27709.799 Da / Num. of mol.: 1 / Fragment: UNP residues 329-569 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: MALT1, MLT / Production host: Homo sapiens (human) / Gene: MALT1, MLT / Production host:  References: UniProt: Q9UDY8, Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases |
|---|---|
| #2: Protein/peptide | 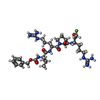  Type: Oligopeptide / Class: Inhibitor / Mass: 697.244 Da / Num. of mol.: 1 / Source method: obtained synthetically Type: Oligopeptide / Class: Inhibitor / Mass: 697.244 Da / Num. of mol.: 1 / Source method: obtained syntheticallyReferences:  PDB-3v4l, N-[(benzyloxy)carbonyl]-L-valyl-N~5~-[amino(iminio)methyl]-L-ornithyl-N-[(3R)-6-{[amino(iminio)methyl]amino}-1-fluoro-2-oxohexan-3-yl]-L-prolinamide PDB-3v4l, N-[(benzyloxy)carbonyl]-L-valyl-N~5~-[amino(iminio)methyl]-L-ornithyl-N-[(3R)-6-{[amino(iminio)methyl]amino}-1-fluoro-2-oxohexan-3-yl]-L-prolinamide |
| #3: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.76 Å3/Da / Density % sol: 55.4 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion / pH: 7 Details: 20% PeG3550, 0.4 Ammonium nitrate, 0.1M Bis-tris propane, VAPOR DIFFUSION, temperature 298K, pH 7.0 |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X10SA / Wavelength: 1 / Wavelength: 1 Å / Beamline: X10SA / Wavelength: 1 / Wavelength: 1 Å | |||||||||
| Detector | Type: PSI PILATUS 6M / Detector: PIXEL / Date: Apr 26, 2010 | |||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||
| Radiation wavelength |
| |||||||||
| Reflection | Resolution: 2.1→67.11 Å / Num. obs: 19456 / % possible obs: 99.7 % / Observed criterion σ(I): -3 / Redundancy: 5.2 % / Rmerge(I) obs: 0.095 / Net I/σ(I): 13.2 | |||||||||
| Reflection shell | Resolution: 2.1→2.15 Å / Redundancy: 5.3 % / Rmerge(I) obs: 0.536 / Mean I/σ(I) obs: 3.6 / % possible all: 99.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 2PYO Resolution: 2.1→53.86 Å / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→53.86 Å
|
 Movie
Movie Controller
Controller












 PDBj
PDBj









