+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3ubp | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DIAMIDOPHOSPHATE INHIBITED BACILLUS PASTEURII UREASE | |||||||||
 Components Components | (PROTEIN (UREASE ...) x 3 | |||||||||
 Keywords Keywords | HYDROLASE / UREASE / BACILLUS PASTEURII / NICKEL / DIAMIDOPHOSPHATE / METALLOENZYME | |||||||||
| Function / homology |  Function and homology information Function and homology informationurease complex / urease / urease activity / urea catabolic process / nickel cation binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Sporosarcina pasteurii (bacteria) Sporosarcina pasteurii (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | |||||||||
 Authors Authors | Benini, S. / Rypniewski, W.R. / Wilson, K.S. / Miletti, S. / Mangani, S. / Ciurli, S. | |||||||||
 Citation Citation |  Journal: Structure Fold.Des. / Year: 1999 Journal: Structure Fold.Des. / Year: 1999Title: A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels. Authors: Benini, S. / Rypniewski, W.R. / Wilson, K.S. / Miletti, S. / Ciurli, S. / Mangani, S. #1:  Journal: Acta Crystallogr.,Sect.D / Year: 1998 Journal: Acta Crystallogr.,Sect.D / Year: 1998Title: Crystallization and Preliminary High-Resolution X-Ray Diffraction Analysis of Native and Beta-Mercaptoethanol-Inhibited Urease from Bacillus Pasteurii Authors: Benini, S. / Ciurli, S. / Rypniewski, W.R. / Wilson, K.S. / Mangani, S. #2:  Journal: J.Biol.Inorg.Chem. / Year: 1998 Journal: J.Biol.Inorg.Chem. / Year: 1998Title: The Complex of Bacillus Pasteurii Urease with Beta-Mercaptoethanol from X-Ray Data at 1.65A Resolution Authors: Benini, S. / Rypniewski, W.R. / Wilson, K.S. / Ciurli, S. / Mangani, S. #3:  Journal: Soil Biol.Biochem. / Year: 1996 Journal: Soil Biol.Biochem. / Year: 1996Title: Bacillus Pasteurii Urease: A Heteropolimeric Enzyme with a Binuclear Nickel Active Site Authors: Benini, S. / Gessa, C. / Ciurli, S. #4:  Journal: Eur.J.Biochem. / Year: 1996 Journal: Eur.J.Biochem. / Year: 1996Title: X-Ray Absorption Spectroscopy Study of Native and Phenylphosphorodiamidate- Inhibited Bacillus Pasteurii Urease Authors: Benini, S. / Ciurli, S. / Nolting, H.F. / Mangani, S. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3ubp.cif.gz 3ubp.cif.gz | 187.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3ubp.ent.gz pdb3ubp.ent.gz | 143.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3ubp.json.gz 3ubp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3ubp_validation.pdf.gz 3ubp_validation.pdf.gz | 451.6 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3ubp_full_validation.pdf.gz 3ubp_full_validation.pdf.gz | 461.8 KB | Display | |
| Data in XML |  3ubp_validation.xml.gz 3ubp_validation.xml.gz | 38.9 KB | Display | |
| Data in CIF |  3ubp_validation.cif.gz 3ubp_validation.cif.gz | 60 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ub/3ubp https://data.pdbj.org/pub/pdb/validation_reports/ub/3ubp ftp://data.pdbj.org/pub/pdb/validation_reports/ub/3ubp ftp://data.pdbj.org/pub/pdb/validation_reports/ub/3ubp | HTTPS FTP |
-Related structure data
| Related structure data |  2ubpSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-PROTEIN (UREASE ... , 3 types, 3 molecules ABC
| #1: Protein | Mass: 11187.017 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Sporosarcina pasteurii (bacteria) / Cellular location: CYTOPLASM / Strain: DSM 33 / References: UniProt: P41022, urease Sporosarcina pasteurii (bacteria) / Cellular location: CYTOPLASM / Strain: DSM 33 / References: UniProt: P41022, urease |
|---|---|
| #2: Protein | Mass: 13975.539 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Sporosarcina pasteurii (bacteria) / Cellular location: CYTOPLASM / Strain: DSM 33 / References: UniProt: P41021, urease Sporosarcina pasteurii (bacteria) / Cellular location: CYTOPLASM / Strain: DSM 33 / References: UniProt: P41021, urease |
| #3: Protein | Mass: 61646.766 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Sporosarcina pasteurii (bacteria) / Cellular location: CYTOPLASM / Strain: DSM 33 / References: UniProt: P41020, urease Sporosarcina pasteurii (bacteria) / Cellular location: CYTOPLASM / Strain: DSM 33 / References: UniProt: P41020, urease |
-Non-polymers , 3 types, 844 molecules 
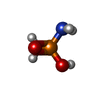



| #4: Chemical | | #5: Chemical | ChemComp-2PA / | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.7 Å3/Da / Density % sol: 54 % | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 6.3 Details: WELL SOLUTIONS: 1.9 M AMMONIUM SULPHATE, 4MM PHENYLPHOSPHORODIAMIDATE, 1OOMM SODIUM CITRATE PH 6.3. PROTEIN SOLUTION: 20 C, 3 MICROLITERS PROTEIN SOLUTION ( 11 MG/ML IN 20 MM TRIS HCL PH 8.0 ...Details: WELL SOLUTIONS: 1.9 M AMMONIUM SULPHATE, 4MM PHENYLPHOSPHORODIAMIDATE, 1OOMM SODIUM CITRATE PH 6.3. PROTEIN SOLUTION: 20 C, 3 MICROLITERS PROTEIN SOLUTION ( 11 MG/ML IN 20 MM TRIS HCL PH 8.0 + 4MM PHENYLPHOSPHORODIAMIDATE) + 3 MICROLITERS PRECIPITANT SOLUTION, VAPOR DIFFUSION, HANGING DROP | |||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS | |||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: BW7A / Wavelength: 0.9995 / Beamline: BW7A / Wavelength: 0.9995 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Jul 24, 1997 / Details: BENT MIRROR |
| Radiation | Monochromator: SI(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9995 Å / Relative weight: 1 |
| Reflection | Resolution: 2→20 Å / Num. obs: 65301 / % possible obs: 99.9 % / Observed criterion σ(I): -3 / Redundancy: 13.38 % / Rmerge(I) obs: 0.15 / Rsym value: 0.15 / Net I/σ(I): 9.72 |
| Reflection shell | Resolution: 2→2.04 Å / Redundancy: 7.56 % / Rmerge(I) obs: 0.536 / Mean I/σ(I) obs: 2.8 / Rsym value: 0.536 / % possible all: 99.9 |
| Reflection | *PLUS Num. measured all: 874166 |
| Reflection shell | *PLUS % possible obs: 99.9 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2UBP Resolution: 2→18 Å / Cross valid method: RFREE / σ(F): 0 / ESU R: 0.13 / ESU R Free: 0.13
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 15.87 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→18 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj



