[English] 日本語
 Yorodumi
Yorodumi- PDB-3qm1: CRYSTAL STRUCTURE OF THE LACTOBACILLUS JOHNSONII CINNAMOYL ESTERA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3qm1 | ||||||
|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURE OF THE LACTOBACILLUS JOHNSONII CINNAMOYL ESTERASE LJ0536 S106A MUTANT IN COMPLEX WITH ETHYLFERULATE, Form II | ||||||
 Components Components | Cinnamoyl esterase | ||||||
 Keywords Keywords | HYDROLASE / ALPHA/BETA HYDROLASE FOLD / CINNAMOYL/FERULOYL ESTERASE / HYDROXYCINAMMATES | ||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on ester bonds; Carboxylic-ester hydrolases / carboxylic ester hydrolase activity Similarity search - Function | ||||||
| Biological species |  Lactobacillus johnsonii (bacteria) Lactobacillus johnsonii (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.817 Å MOLECULAR REPLACEMENT / Resolution: 1.817 Å | ||||||
 Authors Authors | Stogios, P.J. / Lai, K.K. / Vu, C. / Xu, X. / Cui, H. / Molloy, S. / Gonzalez, C.F. / Yakunin, A. / Savchenko, A. | ||||||
 Citation Citation |  Journal: Plos One / Year: 2011 Journal: Plos One / Year: 2011Title: An Inserted alpha/beta Subdomain Shapes the Catalytic Pocket of Lactobacillus johnsonii Cinnamoyl Esterase Authors: Lai, K.K. / Stogios, P.J. / Vu, C. / Xu, X. / Cui, H. / Molloy, S. / Savchenko, A. / Yakunin, A. / Gonzalez, C.F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3qm1.cif.gz 3qm1.cif.gz | 123.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3qm1.ent.gz pdb3qm1.ent.gz | 93.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3qm1.json.gz 3qm1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qm/3qm1 https://data.pdbj.org/pub/pdb/validation_reports/qm/3qm1 ftp://data.pdbj.org/pub/pdb/validation_reports/qm/3qm1 ftp://data.pdbj.org/pub/pdb/validation_reports/qm/3qm1 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3pf8SC  3pf9C  3pfbC  3pfcC  3s2zC  3pfa S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||
| 2 |
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 29449.943 Da / Num. of mol.: 1 / Fragment: UNP residues 22-265 / Mutation: S106A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Lactobacillus johnsonii (bacteria) / Gene: LJ0536 / Plasmid: P15TV-L / Production host: Lactobacillus johnsonii (bacteria) / Gene: LJ0536 / Plasmid: P15TV-L / Production host:  References: UniProt: D3YEX6, Hydrolases; Acting on ester bonds; Carboxylic-ester hydrolases | ||||||
|---|---|---|---|---|---|---|---|
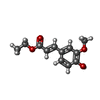
| #2: Chemical | ChemComp-ZYC / | ||||||
| #3: Chemical | ChemComp-NA / #4: Chemical | ChemComp-CL / #5: Water | ChemComp-HOH / | Nonpolymer details | THE BOND ANGLE OAL-CAM-OAC, WITH A VALUE OF 116 DEGREES, IS MODELED IN UNAMBIGUOUS ELECTRON DENSITY ...THE BOND ANGLE OAL-CAM-OAC, WITH A VALUE OF 116 DEGREES, IS MODELED IN UNAMBIGUOU | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.11 Å3/Da / Density % sol: 41.81 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 8.5 Details: 0.1 M Tris pH 8.5, 0.2 M ammonium sulphate, 24% PEG3350, 25 mM ethyl ferulate, 1/10 V8 protease, VAPOR DIFFUSION, SITTING DROP, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU FR-E SUPERBRIGHT / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU FR-E SUPERBRIGHT / Wavelength: 1.5418 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RIGAKU RAXIS HTC / Detector: IMAGE PLATE / Date: Dec 8, 2010 / Details: mirrors | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: mirrors / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.82→23.8 Å / Num. all: 22853 / Num. obs: 22853 / % possible obs: 100 % / Observed criterion σ(F): 2 / Redundancy: 6.1 % / Rsym value: 0.048 / Net I/σ(I): 27.56 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB code 3PF8 Resolution: 1.817→23.067 Å / SU ML: 0.19 / σ(F): 0 / Phase error: 16.72 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 42.33 Å2 / ksol: 0.382 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.817→23.067 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj