[English] 日本語
 Yorodumi
Yorodumi- PDB-3q8j: Crystal Structure of Asteropsin A from Marine Sponge Asteropus sp. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3q8j | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of Asteropsin A from Marine Sponge Asteropus sp. | |||||||||
 Components Components | Asteropsin A | |||||||||
 Keywords Keywords | TOXIN / cystine knot / marine sponge / MARINE KNOTTIN | |||||||||
| Function / homology | METHANOL / Asteropsin A Function and homology information Function and homology information | |||||||||
| Biological species |  Asteropus (invertebrata) Asteropus (invertebrata) | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 0.87 Å X-RAY DIFFRACTION / Resolution: 0.87 Å | |||||||||
 Authors Authors | Bowling, J.J. / Fronczek, F.R. / Hamann, M.T. / Li, H. / Jung, J.H. | |||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: An Uncommon Crystal Structure of a Marine Knottin Peptide from Asteropus sp. Authors: Li, H. / Bowling, J.J. / Fronczek, F.R. / Hong, J. / Hamann, M.T. / Jung, J.H. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3q8j.cif.gz 3q8j.cif.gz | 30.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3q8j.ent.gz pdb3q8j.ent.gz | 20 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3q8j.json.gz 3q8j.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/q8/3q8j https://data.pdbj.org/pub/pdb/validation_reports/q8/3q8j ftp://data.pdbj.org/pub/pdb/validation_reports/q8/3q8j ftp://data.pdbj.org/pub/pdb/validation_reports/q8/3q8j | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 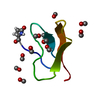
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein/peptide | Mass: 3904.384 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: whole extract / Source: (natural)  Asteropus (invertebrata) / References: UniProt: I1SB10 Asteropus (invertebrata) / References: UniProt: I1SB10 | ||||
|---|---|---|---|---|---|
| #2: Chemical | ChemComp-MOH / #3: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.5634 Å3/Da / Density % sol: 21.3237 % |
|---|---|
| Crystal grow | Temperature: 295 K / Details: CD3OH-H2O, NMR tube, temperature 295K |
-Data collection
| Diffraction | Mean temperature: 90 K |
|---|---|
| Diffraction source | Source: SEALED TUBE / Type: OTHER / Wavelength: 1.54178 Å |
| Detector | Type: APEX II CCD / Detector: CCD / Date: Nov 5, 2010 / Details: MiraCol capillary optic |
| Radiation | Monochromator: Graphite (002) / Protocol: \f and \w scans / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54178 Å / Relative weight: 1 |
| Reflection | Resolution: 0.87→17.15 Å / Num. obs: 27756 / % possible obs: 97.7 % / Redundancy: 2.41 % / Rmerge(I) obs: 0.048 / Net I/σ(I): 11.68 |
| Reflection shell | Resolution: 0.87→0.97 Å / Redundancy: 1 % / Rmerge(I) obs: 0.157 / Mean I/σ(I) obs: 4.4 / % possible all: 60.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 0.87→17.15 Å / Num. parameters: 2725 / Num. restraintsaints: 6 / Occupancy max: 1 / Occupancy min: 1 / Cross valid method: FREE R / σ(F): 0 / Stereochemistry target values: Engh & Huber Details: 1. Patterson to locate S atoms; 2. The sf file contains Friedel pairs.
| |||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 6.2214 Å2 | |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 0 / Occupancy sum hydrogen: 238 / Occupancy sum non hydrogen: 302 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 0.87→17.15 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj