+ Open data
Open data
- Basic information
Basic information
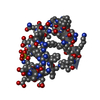
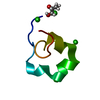
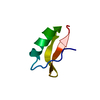
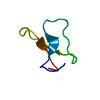
| Entry | Database: PDB / ID: 1ajj | ||||||
|---|---|---|---|---|---|---|---|
| Title | LDL RECEPTOR LIGAND-BINDING MODULE 5, CALCIUM-COORDINATING | ||||||
 Components Components | LOW-DENSITY LIPOPROTEIN RECEPTOR | ||||||
 Keywords Keywords | RECEPTOR / LDL RECEPTOR / CYSTEINE-RICH MODULE / CALCIUM | ||||||
| Function / homology |  Function and homology information Function and homology informationreceptor-mediated endocytosis involved in cholesterol transport / regulation of phosphatidylcholine catabolic process / plasma lipoprotein particle clearance / negative regulation of astrocyte activation / positive regulation of lysosomal protein catabolic process / negative regulation of microglial cell activation / very-low-density lipoprotein particle receptor activity / PCSK9-LDLR complex / cholesterol import / low-density lipoprotein particle clearance ...receptor-mediated endocytosis involved in cholesterol transport / regulation of phosphatidylcholine catabolic process / plasma lipoprotein particle clearance / negative regulation of astrocyte activation / positive regulation of lysosomal protein catabolic process / negative regulation of microglial cell activation / very-low-density lipoprotein particle receptor activity / PCSK9-LDLR complex / cholesterol import / low-density lipoprotein particle clearance / negative regulation of receptor recycling / positive regulation of triglyceride biosynthetic process / clathrin heavy chain binding / intestinal cholesterol absorption / low-density lipoprotein particle receptor activity / Chylomicron clearance / amyloid-beta clearance by cellular catabolic process / low-density lipoprotein particle binding / regulation of protein metabolic process / LDL clearance / high-density lipoprotein particle clearance / response to caloric restriction / lipoprotein catabolic process / phospholipid transport / low-density lipoprotein particle / cholesterol transport / negative regulation of amyloid fibril formation / cellular response to fatty acid / endolysosome membrane / negative regulation of low-density lipoprotein particle clearance / regulation of cholesterol metabolic process / artery morphogenesis / negative regulation of protein metabolic process / sorting endosome / lipoprotein particle binding / amyloid-beta clearance / cellular response to low-density lipoprotein particle stimulus / long-term memory / phagocytosis / retinoid metabolic process / Retinoid metabolism and transport / cholesterol metabolic process / somatodendritic compartment / clathrin-coated pit / receptor-mediated endocytosis / cholesterol homeostasis / lipid metabolic process / clathrin-coated endocytic vesicle membrane / endocytosis / apical part of cell / positive regulation of inflammatory response / late endosome / Cargo recognition for clathrin-mediated endocytosis / amyloid-beta binding / Clathrin-mediated endocytosis / virus receptor activity / protease binding / basolateral plasma membrane / molecular adaptor activity / early endosome / lysosome / receptor complex / endosome membrane / intracellular membrane-bounded organelle / negative regulation of gene expression / external side of plasma membrane / calcium ion binding / positive regulation of gene expression / cell surface / Golgi apparatus / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MIR / Resolution: 1.7 Å MIR / Resolution: 1.7 Å | ||||||
 Authors Authors | Fass, D. / Blacklow, S.C. / Kim, P.S. / Berger, J.M. | ||||||
 Citation Citation |  Journal: Nature / Year: 1997 Journal: Nature / Year: 1997Title: Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Authors: Fass, D. / Blacklow, S. / Kim, P.S. / Berger, J.M. #1:  Journal: Nat.Struct.Biol. / Year: 1996 Journal: Nat.Struct.Biol. / Year: 1996Title: Protein Folding and Calcium Binding Defects Arising from Familial Hypercholesterolemia Mutations of the Ldl Receptor Authors: Blacklow, S.C. / Kim, P.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ajj.cif.gz 1ajj.cif.gz | 20 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ajj.ent.gz pdb1ajj.ent.gz | 11.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ajj.json.gz 1ajj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/aj/1ajj https://data.pdbj.org/pub/pdb/validation_reports/aj/1ajj ftp://data.pdbj.org/pub/pdb/validation_reports/aj/1ajj ftp://data.pdbj.org/pub/pdb/validation_reports/aj/1ajj | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein/peptide | Mass: 4069.431 Da / Num. of mol.: 1 / Fragment: LIGAND-BINDING DOMAIN, FIFTH REPEAT Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Plasmid: PMM-LR5 / Species (production host): Escherichia coli / Production host: Homo sapiens (human) / Plasmid: PMM-LR5 / Species (production host): Escherichia coli / Production host:  |
|---|---|
| #2: Chemical | ChemComp-SO4 / |
| #3: Chemical | ChemComp-CA / |
| #4: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.81 Å3/Da / Density % sol: 28 % | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 5 Details: PROTEIN WAS CRYSTALLIZED FROM 2.1 M AMMONIUM SULFATE, PH 5.0, 25% SUCROSE | |||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 15 ℃ / Method: vapor diffusion, hanging drop | |||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 93 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: RIGAKU / Detector: IMAGE PLATE / Date: Nov 1, 1996 / Details: MIRRORS |
| Radiation | Monochromator: NI FILTER / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.7→15 Å / Num. obs: 2959 / % possible obs: 94.3 % / Observed criterion σ(I): 1.5 / Redundancy: 4.5 % / Rsym value: 0.054 / Net I/σ(I): 20 |
| Reflection shell | Resolution: 1.7→1.76 Å / Redundancy: 3 % / Mean I/σ(I) obs: 7 / Rsym value: 0.169 / % possible all: 87.7 |
| Reflection | *PLUS Rmerge(I) obs: 0.054 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MIR / Resolution: 1.7→15 Å / Cross valid method: THROUGHOUT MIR / Resolution: 1.7→15 Å / Cross valid method: THROUGHOUT
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.7→15 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor Rfree: 0.24 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller









 PDBj
PDBj
















