+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3l2x | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of Spin Labeled T4 Lysozyme Mutant 115-119RX | ||||||
 Components Components | Lysozyme | ||||||
 Keywords Keywords | HYDROLASE / NITROXIDE SPIN LABEL / EPR / MODIFIED CYSTEINE / Antimicrobial / Bacteriolytic enzyme / Glycosidase | ||||||
| Function / homology |  Function and homology information Function and homology informationviral release from host cell by cytolysis / peptidoglycan catabolic process / cell wall macromolecule catabolic process / lysozyme / lysozyme activity / host cell cytoplasm / defense response to bacterium Similarity search - Function | ||||||
| Biological species |  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | ||||||
 Authors Authors | Fleissner, M.R. / Cascio, D. / Hubbell, W.L. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: Crystal Structure of Spin Labeled T4 Lysozyme Mutant 115-119RX Authors: Fleissner, M.R. / Cascio, D. / Kalai, T. / Hideg, K. / Hubbell, W.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3l2x.cif.gz 3l2x.cif.gz | 86.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3l2x.ent.gz pdb3l2x.ent.gz | 63.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3l2x.json.gz 3l2x.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/l2/3l2x https://data.pdbj.org/pub/pdb/validation_reports/l2/3l2x ftp://data.pdbj.org/pub/pdb/validation_reports/l2/3l2x ftp://data.pdbj.org/pub/pdb/validation_reports/l2/3l2x | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1c6tS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 18576.350 Da / Num. of mol.: 1 / Mutation: C54T, C97A, T115C, R119C Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Enterobacteria phage T4 (virus) / Gene: E, Lysozyme / Plasmid: pHSe5 / Production host: Enterobacteria phage T4 (virus) / Gene: E, Lysozyme / Plasmid: pHSe5 / Production host:  |
|---|
-Non-polymers , 6 types, 164 molecules 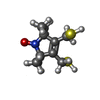









| #2: Chemical | ChemComp-RXR / [ | ||||||
|---|---|---|---|---|---|---|---|
| #3: Chemical | ChemComp-AZI / | ||||||
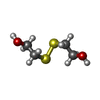
| #4: Chemical | | #5: Chemical | ChemComp-HED / | #6: Chemical | ChemComp-BME / | #7: Water | ChemComp-HOH / | |
-Details
| Nonpolymer details | EXTENSIVE EPR EXPERIMENTS OF THE DISULFIDE LINKED RXR LIGAND ATTACHED TO T4 LYSOZYME AT DIFFERENT ...EXTENSIVE EPR EXPERIMENT |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.71 Å3/Da / Density % sol: 54.68 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 6.4 Details: 2.0M dibasic potassium phosphate and monobasic sodium phosphate, 0.25M sodium chloride, 0.04% sodium azide, saturated with bis(2-hydroxyethyl) disulfide, pH 6.4, vapor diffusion, hanging drop, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU FR-D / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU FR-D / Wavelength: 1.5418 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: Nov 3, 2004 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: CONFOCAL MIRRORS (BLUE OPTICS) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.8→80 Å / Num. obs: 18829 / % possible obs: 97.4 % / Observed criterion σ(F): -3 / Observed criterion σ(I): -3 / Redundancy: 9.3 % / Rsym value: 0.072 / Χ2: 1.023 / Net I/σ(I): 12.3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 1C6T Resolution: 1.8→35.419 Å / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.868 / SU ML: 0.21 / Cross valid method: THROUGHOUT / σ(F): 1.37 / Stereochemistry target values: ML Details: THE RXR LIGAND IS DISUFLDE BONDED TO CYS 115 AND CYS 119, AND WAS MODELED IN AT AN OCCUPANCY OF 0.7
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 40.001 Å2 / ksol: 0.4 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 76.33 Å2 / Biso mean: 21.209 Å2 / Biso min: 8.09 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→35.419 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 7
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 25.2491 Å / Origin y: -11.2388 Å / Origin z: 9.4138 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller























 PDBj
PDBj











