[English] 日本語
 Yorodumi
Yorodumi- PDB-3knc: Crystal structure of the CeNA-RNA hybrid octamer ce(GCGTAGCG):r(C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3knc | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the CeNA-RNA hybrid octamer ce(GCGTAGCG):r(CGCUACGC) | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA/CYCLOHEXENE-RNA HYBRID / cyclohexene / sugar modification / RNA / RNA-CYCLOHEXENE-RNA HYBRID complex | ||||||
| Function / homology | DNA / RNA Function and homology information Function and homology information | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.5 Å molecular replacement / Resolution: 2.5 Å | ||||||
 Authors Authors | Ovaere, M. / Van Meervelt, L. | ||||||
 Citation Citation |  Journal: Chemistry / Year: 2011 Journal: Chemistry / Year: 2011Title: The Crystal Structure of the CeNA:RNA Hybrid ce(GCGTAGCG):r(CGCUACGC). Authors: Ovaere, M. / Herdewijn, P. / Van Meervelt, L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3knc.cif.gz 3knc.cif.gz | 23.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3knc.ent.gz pdb3knc.ent.gz | 14.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3knc.json.gz 3knc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3knc_validation.pdf.gz 3knc_validation.pdf.gz | 403.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3knc_full_validation.pdf.gz 3knc_full_validation.pdf.gz | 406 KB | Display | |
| Data in XML |  3knc_validation.xml.gz 3knc_validation.xml.gz | 4.5 KB | Display | |
| Data in CIF |  3knc_validation.cif.gz 3knc_validation.cif.gz | 5.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kn/3knc https://data.pdbj.org/pub/pdb/validation_reports/kn/3knc ftp://data.pdbj.org/pub/pdb/validation_reports/kn/3knc ftp://data.pdbj.org/pub/pdb/validation_reports/kn/3knc | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 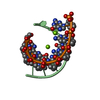
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
| ||||||||
| Details | Biological unit is the same as asymmetric unit |
- Components
Components
| #1: DNA chain | Mass: 2887.177 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: The CeNA strand was synthesized. | ||
|---|---|---|---|
| #2: RNA chain | Mass: 2501.553 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: The RNA strand was synthesized. | ||
| #3: Chemical | | #4: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.22 Å3/Da / Density % sol: 61.82 % Description: The structure factor file contains Friedel pairs | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 289 K / Method: vapor diffusion, hanging drop Details: 0.2M Magnesium formate, VAPOR DIFFUSION, HANGING DROP, temperature 289K | ||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 1 Å / Beamline: X06DA / Wavelength: 1 Å |
| Detector | Type: MARMOSAIC 225 mm CCD / Detector: CCD / Date: Sep 29, 2008 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.5→36.3 Å / Num. all: 2460 / Num. obs: 2575 / % possible obs: 98.8 % / Redundancy: 9.4 % / Rsym value: 0.048 |
-Phasing
| Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR | Rfactor: 31.37 / Model details: Phaser MODE: MR_AUTO
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.5→36.3 Å / Cor.coef. Fo:Fc: 0.97 / Cor.coef. Fo:Fc free: 0.958 / WRfactor Rfree: 0.232 / WRfactor Rwork: 0.184 / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.762 / SU B: 9.838 / SU ML: 0.19 / SU R Cruickshank DPI: 0.31 / SU Rfree: 0.237 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.31 / ESU R Free: 0.237 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: The Friedel pairs were used in phasing MOLECULAR REPLACEMENT / Resolution: 2.5→36.3 Å / Cor.coef. Fo:Fc: 0.97 / Cor.coef. Fo:Fc free: 0.958 / WRfactor Rfree: 0.232 / WRfactor Rwork: 0.184 / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.762 / SU B: 9.838 / SU ML: 0.19 / SU R Cruickshank DPI: 0.31 / SU Rfree: 0.237 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.31 / ESU R Free: 0.237 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: The Friedel pairs were used in phasing
| ||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 83.75 Å2 / Biso mean: 61.046 Å2 / Biso min: 37.01 Å2
| ||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.5→36.3 Å
| ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.5→2.57 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller







 PDBj
PDBj


