[English] 日本語
 Yorodumi
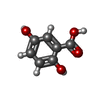
Yorodumi- PDB-3jut: Acidic Fibroblast Growth Factor (FGF-1) complexed with gentisic acid -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3jut | ||||||
|---|---|---|---|---|---|---|---|
| Title | Acidic Fibroblast Growth Factor (FGF-1) complexed with gentisic acid | ||||||
 Components Components | Heparin-binding growth factor 1 | ||||||
 Keywords Keywords | HORMONE / GROWTH FACTOR / FGF-1 INHIBITORS / Angiogenesis / Developmental protein / Differentiation / Heparin-binding / Mitogen | ||||||
| Function / homology |  Function and homology information Function and homology informationmesonephric epithelium development / branch elongation involved in ureteric bud branching / regulation of endothelial tube morphogenesis / FGFR3b ligand binding and activation / regulation of endothelial cell chemotaxis to fibroblast growth factor / Signaling by activated point mutants of FGFR3 / FGFR3c ligand binding and activation / Phospholipase C-mediated cascade; FGFR3 / FGFR2b ligand binding and activation / fibroblast growth factor receptor binding ...mesonephric epithelium development / branch elongation involved in ureteric bud branching / regulation of endothelial tube morphogenesis / FGFR3b ligand binding and activation / regulation of endothelial cell chemotaxis to fibroblast growth factor / Signaling by activated point mutants of FGFR3 / FGFR3c ligand binding and activation / Phospholipase C-mediated cascade; FGFR3 / FGFR2b ligand binding and activation / fibroblast growth factor receptor binding / FGFR2c ligand binding and activation / Activated point mutants of FGFR2 / FGFR4 ligand binding and activation / Phospholipase C-mediated cascade; FGFR2 / FGFR1b ligand binding and activation / Phospholipase C-mediated cascade; FGFR4 / Signaling by activated point mutants of FGFR1 / FGFR1c ligand binding and activation / organ induction / Downstream signaling of activated FGFR1 / Phospholipase C-mediated cascade: FGFR1 / S100 protein binding / activation of protein kinase B activity / Signaling by FGFR2 IIIa TM / PI-3K cascade:FGFR3 / PI-3K cascade:FGFR2 / PI-3K cascade:FGFR4 / PI-3K cascade:FGFR1 / positive regulation of sprouting angiogenesis / positive regulation of MAP kinase activity / positive regulation of cell division / positive regulation of intracellular signal transduction / PI3K Cascade / fibroblast growth factor receptor signaling pathway / anatomical structure morphogenesis / SHC-mediated cascade:FGFR3 / SHC-mediated cascade:FGFR2 / SHC-mediated cascade:FGFR4 / SHC-mediated cascade:FGFR1 / FRS-mediated FGFR3 signaling / FRS-mediated FGFR2 signaling / FRS-mediated FGFR4 signaling / FRS-mediated FGFR1 signaling / Signaling by FGFR3 in disease / Signaling by FGFR2 in disease / neurogenesis / Signaling by FGFR1 in disease / positive regulation of endothelial cell migration / lung development / regulation of cell migration / epithelial cell proliferation / positive regulation of epithelial cell proliferation / growth factor activity / Negative regulation of FGFR3 signaling / Negative regulation of FGFR2 signaling / wound healing / Negative regulation of FGFR4 signaling / Negative regulation of FGFR1 signaling / positive regulation of cholesterol biosynthetic process / integrin binding / extracellular matrix / Constitutive Signaling by Aberrant PI3K in Cancer / positive regulation of angiogenesis / PIP3 activates AKT signaling / heparin binding / cellular response to heat / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / RAF/MAP kinase cascade / angiogenesis / cell cortex / positive regulation of ERK1 and ERK2 cascade / positive regulation of MAPK cascade / positive regulation of cell migration / positive regulation of cell population proliferation / signal transduction / positive regulation of transcription by RNA polymerase II / extracellular space / extracellular region / nucleoplasm / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.25 Å MOLECULAR REPLACEMENT / Resolution: 2.25 Å | ||||||
 Authors Authors | Fernandez, I.S. / Gimenez-Gallego, G. / Romero, A. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2010 Journal: J.Biol.Chem. / Year: 2010Title: Gentisic acid, a compound associated with plant defense and a metabolite of aspirin, heads a new class of in vivo fibroblast growth factor inhibitors. Authors: Fernandez, I.S. / Cuevas, P. / Angulo, J. / Lopez-Navajas, P. / Canales-Mayordomo, A. / Gonzalez-Corrochano, R. / Lozano, R.M. / Valverde, S. / Jimenez-Barbero, J. / Romero, A. / Gimenez-Gallego, G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3jut.cif.gz 3jut.cif.gz | 162 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3jut.ent.gz pdb3jut.ent.gz | 130.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3jut.json.gz 3jut.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ju/3jut https://data.pdbj.org/pub/pdb/validation_reports/ju/3jut ftp://data.pdbj.org/pub/pdb/validation_reports/ju/3jut ftp://data.pdbj.org/pub/pdb/validation_reports/ju/3jut | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3k1xC  1axmS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 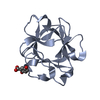
| ||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||||||||||||
| 3 | 
| ||||||||||||||||||||||||||||||||||||||||||
| 4 | 
| ||||||||||||||||||||||||||||||||||||||||||
| 5 | 
| ||||||||||||||||||||||||||||||||||||||||||
| 6 | 
| ||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: 1 / Beg auth comp-ID: LYS / Beg label comp-ID: LYS / End auth comp-ID: SER / End label comp-ID: SER / Refine code: 6 / Auth seq-ID: 1 - 130 / Label seq-ID: 1 - 130
|
- Components
Components
| #1: Protein | Mass: 14752.703 Da / Num. of mol.: 6 / Fragment: Heparin-binding, UNP residues 24-153 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Plasmid: pRAT-4 / Production host: Homo sapiens (human) / Plasmid: pRAT-4 / Production host:  #2: Chemical | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.48 Å3/Da / Density % sol: 50.46 % |
|---|---|
| Crystal grow | Temperature: 295 K / Method: vapor diffusion, sitting drop / pH: 7.8 Details: Equal volumes of protein and inhibitor solutions, 0.75 and 1.5 mM,respectively were mixed with drops containing 60% sodium/potassium tartrate buffered with 5 mM sodium phosphate [pH 7.8]. ...Details: Equal volumes of protein and inhibitor solutions, 0.75 and 1.5 mM,respectively were mixed with drops containing 60% sodium/potassium tartrate buffered with 5 mM sodium phosphate [pH 7.8]. The drops were equilibrated against 200 ml of 1.3M Li2SO4 and typical crystals grew within two weeks , VAPOR DIFFUSION, SITTING DROP, temperature 295K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: BM16 / Wavelength: 0.979 Å / Beamline: BM16 / Wavelength: 0.979 Å |
| Detector | Type: ADSC QUANTUM 210r / Detector: CCD / Date: Dec 7, 2008 |
| Radiation | Monochromator: Si 111 CHANNEL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.979 Å / Relative weight: 1 |
| Reflection | Resolution: 2.21→47.7 Å / Num. all: 42342 / Num. obs: 41838 / % possible obs: 98.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3.3 % / Biso Wilson estimate: 31.73 Å2 / Rmerge(I) obs: 0.082 / Net I/σ(I): 5.6 |
| Reflection shell | Resolution: 2.21→2.33 Å / Redundancy: 3.3 % / Rmerge(I) obs: 0.328 / Mean I/σ(I) obs: 1.6 / Num. unique all: 5625 / % possible all: 94.6 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1axm Resolution: 2.25→37.01 Å / Cor.coef. Fo:Fc: 0.943 / Cor.coef. Fo:Fc free: 0.91 / SU B: 17.715 / SU ML: 0.203 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / ESU R: 0.346 / ESU R Free: 0.258 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 38.52 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.25→37.01 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Ens-ID: 1 / Number: 1031 / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.25→2.308 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj