[English] 日本語
 Yorodumi
Yorodumi- PDB-3fma: Crystal structure of the GYF domain of Smy2 in complex with a pro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3fma | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the GYF domain of Smy2 in complex with a proline-rich peptide from BBP/ScSF1 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PROTEIN BINDING / GYF domain / poly-proline binding / proline-rich peptide / domain swap / PRS / RAGNYA | ||||||
| Function / homology |  Function and homology information Function and homology informationpre-mRNA branch point binding / nuclear protein quality control by the ubiquitin-proteasome system / protein-containing complex disassembly / regulation of mRNA splicing, via spliceosome / commitment complex / spliceosomal complex assembly / endoplasmic reticulum to Golgi vesicle-mediated transport / mRNA splicing, via spliceosome / mRNA binding / endoplasmic reticulum membrane ...pre-mRNA branch point binding / nuclear protein quality control by the ubiquitin-proteasome system / protein-containing complex disassembly / regulation of mRNA splicing, via spliceosome / commitment complex / spliceosomal complex assembly / endoplasmic reticulum to Golgi vesicle-mediated transport / mRNA splicing, via spliceosome / mRNA binding / endoplasmic reticulum membrane / regulation of transcription by RNA polymerase II / zinc ion binding / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2.5 Å MAD / Resolution: 2.5 Å | ||||||
 Authors Authors | Ash, M.R. / Faelber, K. | ||||||
 Citation Citation |  Journal: to be published Journal: to be publishedTitle: SMY2-type GYF domain recognition in mRNA surveillance complexes Authors: Ash, M.R. / Faelber, K. / Kosslick, D. / Albert, G. / Roske, Y. / Kofler, M. / Schuemann, M. / Krause, E. / Freund, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3fma.cif.gz 3fma.cif.gz | 99.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3fma.ent.gz pdb3fma.ent.gz | 78.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3fma.json.gz 3fma.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3fma_validation.pdf.gz 3fma_validation.pdf.gz | 488.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3fma_full_validation.pdf.gz 3fma_full_validation.pdf.gz | 491.7 KB | Display | |
| Data in XML |  3fma_validation.xml.gz 3fma_validation.xml.gz | 18 KB | Display | |
| Data in CIF |  3fma_validation.cif.gz 3fma_validation.cif.gz | 24.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fm/3fma https://data.pdbj.org/pub/pdb/validation_reports/fm/3fma ftp://data.pdbj.org/pub/pdb/validation_reports/fm/3fma ftp://data.pdbj.org/pub/pdb/validation_reports/fm/3fma | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 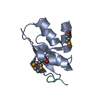
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: 1 / End auth comp-ID: THR / End label comp-ID: THR / Refine code: 4
|
- Components
Components
| #1: Protein | Mass: 11296.908 Da / Num. of mol.: 5 / Fragment: GYF domain Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: SMY2 / Plasmid: pET28 / Production host:  #2: Protein/peptide | Mass: 982.090 Da / Num. of mol.: 5 / Fragment: Proline-rich peptide / Source method: obtained synthetically / Details: Fmoc solid phase synthesis / References: UniProt: Q12186 #3: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.15 Å3/Da / Density % sol: 61.01 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 9 Details: 1.2M (NH4)2SO4, 0.1M Bicine, pH 9.0, vapor diffusion, sitting drop, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: BM14 / Wavelength: 0.978522, 0.97875 / Beamline: BM14 / Wavelength: 0.978522, 0.97875 | |||||||||
| Detector | Type: MARMOSAIC 225 mm CCD / Detector: CCD / Date: Mar 6, 2007 | |||||||||
| Radiation | Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||
| Radiation wavelength |
| |||||||||
| Reflection | Resolution: 2.5→48.057 Å / Num. obs: 27942 / % possible obs: 100 % / Observed criterion σ(I): -3 / Redundancy: 15.1 % / Biso Wilson estimate: 64.4 Å2 / Rmerge(I) obs: 0.123 / Net I/σ(I): 17.8 | |||||||||
| Reflection shell | Resolution: 2.5→2.64 Å / Redundancy: 10.5 % / Rmerge(I) obs: 0.878 / Mean I/σ(I) obs: 2.1 / Num. measured all: 41869 / Num. unique all: 4003 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD / Resolution: 2.5→48.057 Å / Cor.coef. Fo:Fc: 0.94 / Cor.coef. Fo:Fc free: 0.933 / WRfactor Rfree: 0.252 / WRfactor Rwork: 0.22 / Occupancy max: 1 / Occupancy min: 0.3 / FOM work R set: 0.805 / SU B: 22.043 / SU ML: 0.205 / SU R Cruickshank DPI: 0.313 / SU Rfree: 0.235 / TLS residual ADP flag: LIKELY RESIDUAL / Isotropic thermal model: Isotropic with TLS / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.313 / ESU R Free: 0.235 / Stereochemistry target values: MAXIMUM LIKELIHOOD MAD / Resolution: 2.5→48.057 Å / Cor.coef. Fo:Fc: 0.94 / Cor.coef. Fo:Fc free: 0.933 / WRfactor Rfree: 0.252 / WRfactor Rwork: 0.22 / Occupancy max: 1 / Occupancy min: 0.3 / FOM work R set: 0.805 / SU B: 22.043 / SU ML: 0.205 / SU R Cruickshank DPI: 0.313 / SU Rfree: 0.235 / TLS residual ADP flag: LIKELY RESIDUAL / Isotropic thermal model: Isotropic with TLS / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.313 / ESU R Free: 0.235 / Stereochemistry target values: MAXIMUM LIKELIHOODDetails: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS, U VALUES: RESIDUAL ONLY
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 48.84 Å2 / Biso mean: 30.558 Å2 / Biso min: 18.71 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.5→48.057 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Dom-ID: 1 / Ens-ID: 1 / Number: 670 / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.5→2.565 Å / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj



